Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2
- PMID: 33690649
- PMCID: PMC7946320
- DOI: 10.1371/journal.pone.0248348
Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2
Abstract
Pseudoviruses are useful surrogates for highly pathogenic viruses because of their safety, genetic stability, and scalability for screening assays. Many different pseudovirus platforms exist, each with different advantages and limitations. Here we report our efforts to optimize and characterize an HIV-based lentiviral pseudovirus assay for screening neutralizing antibodies for SARS-CoV-2 using a stable 293T cell line expressing human angiotensin converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2). We assessed different target cells, established conditions that generate readouts over at least a two-log range, and confirmed consistent neutralization titers over a range of pseudovirus input. Using reference sera and plasma panels, we evaluated assay precision and showed that our neutralization titers correlate well with results reported in other assays. Overall, our lentiviral assay is relatively simple, scalable, and suitable for a variety of SARS-CoV-2 entry and neutralization screening assays.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
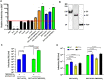

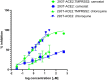
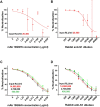
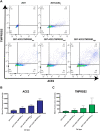

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

