A prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant from benign pulmonary nodules
- PMID: 33691657
- PMCID: PMC7944594
- DOI: 10.1186/s12885-021-08002-4
A prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant from benign pulmonary nodules
Abstract
Background: Lung cancer remains the leading cause of cancer deaths across the world. Early detection of lung cancer by low-dose computed tomography (LDCT) can reduce the mortality rate. However, making a definitive preoperative diagnosis of malignant pulmonary nodules (PNs) found by LDCT is a clinical challenge. This study aimed to develop a prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant pulmonary nodules from benign PNs.
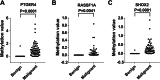
Methods: We assessed three DNA methylation biomarkers (PTGER4, RASSF1A, and SHOX2) and clinically-relevant variables in a training cohort of 110 individuals with PNs. Four machine-learning-based prediction models were established and compared, including the K-nearest neighbors (KNN), random forest (RF), support vector machine (SVM), and logistic regression (LR) algorithms. Variables of the best-performing algorithm (LR) were selected through stepwise use of Akaike's information criterion (AIC). The constructed prediction model was compared with the methylation biomarkers and the Mayo Clinic model using the non-parametric approach of DeLong et al. with the area under a receiver operator characteristic curve (AUC) analysis.
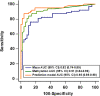
Results: A prediction model was finally constructed based on three DNA methylation biomarkers and one radiological characteristic for identifying malignant from benign PNs. The developed prediction model achieved an AUC value of 0.951 in malignant PNs diagnosis, significantly higher than the three DNA methylation biomarkers (0.912, 95% CI:0.843-0.958, p = 0.013) or Mayo Clinic model (0.823, 95% CI:0.739-0.890, p = 0.001). Validation of the prediction model in the testing cohort of 100 subjects with PNs confirmed the diagnostic value.
Conclusion: We have shown that integrating DNA methylation biomarkers and radiological characteristics could more accurately identify lung cancer in subjects with CT-found PNs. The prediction model developed in our study may provide clinical utility in combination with LDCT to improve the over-all diagnosis of lung cancer.
Keywords: Biomarkers; CT; DNA methylation; Lung cancer; Pulmonary nodules.
Conflict of interest statement
ML is an employee of Excellen Medical Technology Co., Ltd. All other authors have no conflicts of interest to declare.
Figures


Similar articles
-
A combined model of circulating tumor DNA methylated SHOX2/SCT/HOXA7 and clinical features facilitates the discrimination of malignant from benign pulmonary nodules.Lung Cancer. 2025 Jan;199:108064. doi: 10.1016/j.lungcan.2024.108064. Epub 2024 Dec 16. Lung Cancer. 2025. PMID: 39705824
-
DNA methylation of PTGER4 in peripheral blood plasma helps to distinguish between lung cancer, benign pulmonary nodules and chronic obstructive pulmonary disease patients.Eur J Cancer. 2021 Apr;147:142-150. doi: 10.1016/j.ejca.2021.01.032. Epub 2021 Mar 1. Eur J Cancer. 2021. PMID: 33662689
-
Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease.J Thorac Oncol. 2017 Jan;12(1):77-84. doi: 10.1016/j.jtho.2016.08.123. Epub 2016 Aug 18. J Thorac Oncol. 2017. PMID: 27544059 Free PMC article. Clinical Trial.
-
Current advances and future prospects of blood-based techniques for identifying benign and malignant pulmonary nodules.Crit Rev Oncol Hematol. 2025 Mar;207:104608. doi: 10.1016/j.critrevonc.2024.104608. Epub 2025 Jan 4. Crit Rev Oncol Hematol. 2025. PMID: 39761937 Review.
-
Application of liquid biopsy in differentiating lung cancer from benign pulmonary nodules (Review).Int J Mol Med. 2025 Jul;56(1):106. doi: 10.3892/ijmm.2025.5547. Epub 2025 May 9. Int J Mol Med. 2025. PMID: 40341969 Free PMC article. Review.
Cited by
-
A novel multimodal prediction model based on DNA methylation biomarkers and low-dose computed tomography images for identifying early-stage lung cancer.Chin J Cancer Res. 2023 Oct 30;35(5):511-525. doi: 10.21147/j.issn.1000-9604.2023.05.08. Chin J Cancer Res. 2023. PMID: 37969955 Free PMC article.
-
Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer.Pharmaceuticals (Basel). 2023 Jul 22;16(7):1042. doi: 10.3390/ph16071042. Pharmaceuticals (Basel). 2023. PMID: 37513953 Free PMC article. Review.
-
Machine learning-derived peripheral blood transcriptomic biomarkers for early lung cancer diagnosis: Unveiling tumor-immune interaction mechanisms.Biofactors. 2025 Jan-Feb;51(1):e2129. doi: 10.1002/biof.2129. Epub 2024 Oct 16. Biofactors. 2025. PMID: 39415336 Free PMC article.
-
Minimally invasive biomarkers for triaging lung nodules-challenges and future perspectives.Cancer Metastasis Rev. 2025 Jan 31;44(1):29. doi: 10.1007/s10555-025-10247-5. Cancer Metastasis Rev. 2025. PMID: 39888565 Free PMC article. Review.
-
Promising predictive molecular biomarkers for cervical cancer (Review).Int J Mol Med. 2024 Jun;53(6):50. doi: 10.3892/ijmm.2024.5374. Epub 2024 Apr 12. Int J Mol Med. 2024. PMID: 38606495 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

