Pattern-recognition receptors are required for NLR-mediated plant immunity
- PMID: 33692546
- PMCID: PMC8016741
- DOI: 10.1038/s41586-021-03316-6
Pattern-recognition receptors are required for NLR-mediated plant immunity
Abstract
The plant immune system is fundamental for plant survival in natural ecosystems and for productivity in crop fields. Substantial evidence supports the prevailing notion that plants possess a two-tiered innate immune system, called pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is triggered by microbial patterns via cell surface-localized pattern-recognition receptors (PRRs), whereas ETI is activated by pathogen effector proteins via predominantly intracellularly localized receptors called nucleotide-binding, leucine-rich repeat receptors (NLRs)1-4. PTI and ETI are initiated by distinct activation mechanisms and involve different early signalling cascades5,6. Here we show that Arabidopsis PRR and PRR co-receptor mutants-fls2 efr cerk1 and bak1 bkk1 cerk1 triple mutants-are markedly impaired in ETI responses when challenged with incompatible Pseudomonas syrinage bacteria. We further show that the production of reactive oxygen species by the NADPH oxidase RBOHD is a critical early signalling event connecting PRR- and NLR-mediated immunity, and that the receptor-like cytoplasmic kinase BIK1 is necessary for full activation of RBOHD, gene expression and bacterial resistance during ETI. Moreover, NLR signalling rapidly augments the transcript and/or protein levels of key PTI components. Our study supports a revised model in which potentiation of PTI is an indispensable component of ETI during bacterial infection. This revised model conceptually unites two major immune signalling cascades in plants and mechanistically explains some of the long-observed similarities in downstream defence outputs between PTI and ETI.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
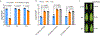

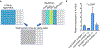








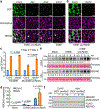


Comment in
-
Plant immunity unified.Nat Plants. 2021 Apr;7(4):382-383. doi: 10.1038/s41477-021-00903-3. Nat Plants. 2021. PMID: 33785867 No abstract available.
-
Plant immunity: Crosstalk between plant immune receptors.Curr Biol. 2021 Jun 21;31(12):R796-R798. doi: 10.1016/j.cub.2021.04.080. Curr Biol. 2021. PMID: 34157265
References
-
- Couto D & Zipfel C Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 16, 537–552 (2016). - PubMed
-
- Spoel SH & Dong X How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 12, 89–100 (2012). - PubMed
-
- Cui H, Tsuda K & Parker JE Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 66, 487–511 (2015). - PubMed
-
- Jones JD & Dangl JL The plant immune system. Nature. 444, 323–329 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

