High Throughput Sequencing for the Detection and Characterization of RNA Viruses
- PMID: 33692767
- PMCID: PMC7938315
- DOI: 10.3389/fmicb.2021.621719
High Throughput Sequencing for the Detection and Characterization of RNA Viruses
Abstract
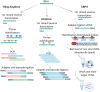
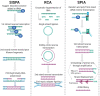
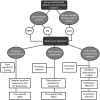
This review aims to assess and recommend approaches for targeted and agnostic High Throughput Sequencing of RNA viruses in a variety of sample matrices. HTS also referred to as deep sequencing, next generation sequencing and third generation sequencing; has much to offer to the field of environmental virology as its increased sequencing depth circumvents issues with cloning environmental isolates for Sanger sequencing. That said however, it is important to consider the challenges and biases that method choice can impart to sequencing results. Here, methodology choices from RNA extraction, reverse transcription to library preparation are compared based on their impact on the detection or characterization of RNA viruses.
Keywords: RNA depletion; RNA viruses; amplicon sequencing; capture based probe hybridization; environmental virology; high throughput sequencing; viral enrichment.
Copyright © 2021 Fitzpatrick, Rupnik, O'Shea, Crispie, Keaveney and Cotter.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ahmed R., Hossain M. S., Haque M. S., Alam M. M., Islam M. S. (2019). Modified protocol for RNA isolation from different parts of field-grown jute plant suitable for NGS data generation and quantitative real-time RT-PCR. Afr. J. Biotechnol. 18, 647–658. 10.5897/AJB2019.16819 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources