An RNAi screen of the kinome in epithelial follicle cells of the Drosophila melanogaster ovary reveals genes required for proper germline death and clearance
- PMID: 33693600
- PMCID: PMC8022946
- DOI: 10.1093/g3journal/jkaa066
An RNAi screen of the kinome in epithelial follicle cells of the Drosophila melanogaster ovary reveals genes required for proper germline death and clearance
Abstract
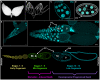
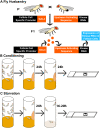
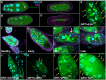
Programmed cell death and cell corpse clearance are an essential part of organismal health and development. Cell corpses are often cleared away by professional phagocytes such as macrophages. However, in certain tissues, neighboring cells known as nonprofessional phagocytes can also carry out clearance functions. Here, we use the Drosophila melanogaster ovary to identify novel genes required for clearance by nonprofessional phagocytes. In the Drosophila ovary, germline cells can die at multiple time points. As death proceeds, the epithelial follicle cells act as phagocytes to facilitate the clearance of these cells. We performed an unbiased kinase screen to identify novel proteins and pathways involved in cell clearance during two death events. Of 224 genes examined, 18 demonstrated severe phenotypes during developmental death and clearance while 12 demonstrated severe phenotypes during starvation-induced cell death and clearance, representing a number of pathways not previously implicated in phagocytosis. Interestingly, it was found that several genes not only affected the clearance process in the phagocytes, but also non-autonomously affected the process by which germline cells died. This kinase screen has revealed new avenues for further exploration and investigation.
Keywords: Drosophila; cell death; kinase; phagocytosis; phosphoinositide 3-kinase.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
