Mechanisms of damage tolerance and repair during DNA replication
- PMID: 33693881
- PMCID: PMC8034635
- DOI: 10.1093/nar/gkab101
Mechanisms of damage tolerance and repair during DNA replication
Abstract
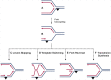
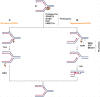
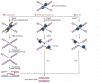
Accurate duplication of chromosomal DNA is essential for the transmission of genetic information. The DNA replication fork encounters template lesions, physical barriers, transcriptional machinery, and topological barriers that challenge the faithful completion of the replication process. The flexibility of replisomes coupled with tolerance and repair mechanisms counteract these replication fork obstacles. The cell possesses several universal mechanisms that may be activated in response to various replication fork impediments, but it has also evolved ways to counter specific obstacles. In this review, we will discuss these general and specific strategies to counteract different forms of replication associated damage to maintain genomic stability.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

