Frustrated folding of guanine quadruplexes in telomeric DNA
- PMID: 33693924
- PMCID: PMC8034632
- DOI: 10.1093/nar/gkab140
Frustrated folding of guanine quadruplexes in telomeric DNA
Abstract
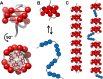
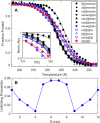
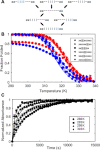
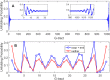
Human chromosomes terminate in long, single-stranded, DNA overhangs of the repetitive sequence (TTAGGG)n. Sets of four adjacent TTAGGG repeats can fold into guanine quadruplexes (GQ), four-stranded structures that are implicated in telomere maintenance and cell immortalization and are targets in cancer therapy. Isolated GQs have been studied in detail, however much less is known about folding in long repeat sequences. Such chains adopt an enormous number of configurations containing various arrangements of GQs and unfolded gaps, leading to a highly frustrated energy landscape. To better understand this phenomenon, we used mutagenesis, thermal melting, and global analysis to determine stability, kinetic, and cooperativity parameters for GQ folding within chains containing 8-12 TTAGGG repeats. We then used these parameters to simulate the folding of 32-repeat chains, more representative of intact telomeres. We found that a combination of folding frustration and negative cooperativity between adjacent GQs increases TTAGGG unfolding by up to 40-fold, providing an abundance of unfolded gaps that are potential binding sites for telomeric proteins. This effect was most pronounced at the chain termini, which could promote telomere extension by telomerase. We conclude that folding frustration is an important and largely overlooked factor controlling the structure of telomeric DNA.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

