MIR503HG Loss Promotes Endothelial-to-Mesenchymal Transition in Vascular Disease
- PMID: 33703914
- PMCID: PMC7610629
- DOI: 10.1161/CIRCRESAHA.120.318124
MIR503HG Loss Promotes Endothelial-to-Mesenchymal Transition in Vascular Disease
Abstract
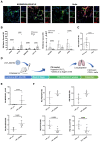
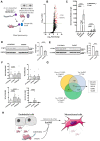
[Figure: see text].
Keywords: endothelial cells; lncRNA; microRNAs; pulmonary arterial hypertension; vascular remodeling.
Figures



References
-
- Manavski Y, Lucas T, Glaser SF, et al. Clonal expansion of endothelial cells contributes to ischemia-induced neovascularization. Circ Res. 2018;122:670–677. - PubMed
-
- Zeisberg EM, Tarnavski O, Zeisberg M, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- SP/17/3/33020/BHF_/British Heart Foundation/United Kingdom
- RG/14/3/30706/BHF_/British Heart Foundation/United Kingdom
- 338991/ERC_/European Research Council/International
- R01 HL148167/HL/NHLBI NIH HHS/United States
- 208402/WT_/Wellcome Trust/United Kingdom
- PG/21/10624/BHF_/British Heart Foundation/United Kingdom
- CH/09/001/25945/BHF_/British Heart Foundation/United Kingdom
- R01 HL135093/HL/NHLBI NIH HHS/United States
- R01 HL130423/HL/NHLBI NIH HHS/United States
- FS/16/38/32351/BHF_/British Heart Foundation/United Kingdom
- CH/11/2/28733/BHF_/British Heart Foundation/United Kingdom
- FS/18/40/33650/BHF_/British Heart Foundation/United Kingdom
- PG/20/10347/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

