Dissecting autism and schizophrenia through neuroimaging genomics
- PMID: 33704401
- PMCID: PMC8370419
- DOI: 10.1093/brain/awab096
Dissecting autism and schizophrenia through neuroimaging genomics
Abstract
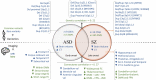
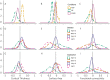
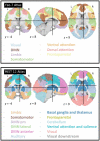
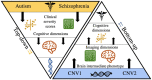
Neuroimaging genomic studies of autism spectrum disorder and schizophrenia have mainly adopted a 'top-down' approach, beginning with the behavioural diagnosis, and moving down to intermediate brain phenotypes and underlying genetic factors. Advances in imaging and genomics have been successfully applied to increasingly large case-control studies. As opposed to diagnostic-first approaches, the bottom-up strategy begins at the level of molecular factors enabling the study of mechanisms related to biological risk, irrespective of diagnoses or clinical manifestations. The latter strategy has emerged from questions raised by top-down studies: why are mutations and brain phenotypes over-represented in individuals with a psychiatric diagnosis? Are they related to core symptoms of the disease or to comorbidities? Why are mutations and brain phenotypes associated with several psychiatric diagnoses? Do they impact a single dimension contributing to all diagnoses? In this review, we aimed at summarizing imaging genomic findings in autism and schizophrenia as well as neuropsychiatric variants associated with these conditions. Top-down studies of autism and schizophrenia identified patterns of neuroimaging alterations with small effect-sizes and an extreme polygenic architecture. Genomic variants and neuroimaging patterns are shared across diagnostic categories suggesting pleiotropic mechanisms at the molecular and brain network levels. Although the field is gaining traction; characterizing increasingly reproducible results, it is unlikely that top-down approaches alone will be able to disentangle mechanisms involved in autism or schizophrenia. In stark contrast with top-down approaches, bottom-up studies showed that the effect-sizes of high-risk neuropsychiatric mutations are equally large for neuroimaging and behavioural traits. Low specificity has been perplexing with studies showing that broad classes of genomic variants affect a similar range of behavioural and cognitive dimensions, which may be consistent with the highly polygenic architecture of psychiatric conditions. The surprisingly discordant effect sizes observed between genetic and diagnostic first approaches underscore the necessity to decompose the heterogeneity hindering case-control studies in idiopathic conditions. We propose a systematic investigation across a broad spectrum of neuropsychiatric variants to identify putative latent dimensions underlying idiopathic conditions. Gene expression data on temporal, spatial and cell type organization in the brain have also considerable potential for parsing the mechanisms contributing to these dimensions' phenotypes. While large neuroimaging genomic datasets are now available in unselected populations, there is an urgent need for data on individuals with a range of psychiatric symptoms and high-risk genomic variants. Such efforts together with more standardized methods will improve mechanistically informed predictive modelling for diagnosis and clinical outcomes.
Keywords: autism; copy number variants; neuroimaging; schizophrenia.
© The Author(s) (2021). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures
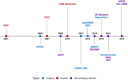
References
-
- Bleuler E.Dementia praecox oder Gruppe der Schizophrenien. Deuticke; 1911.
-
- Kanner L.Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. - PubMed
-
- Asperger H.Die “Autistischen Psychopathen” im Kindesalter. Archiv Für Psychiatrie Und Nervenkrankheiten. 1944;117(1):76–136.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

