Chromosome inheritance and meiotic stability in allopolyploid Brassica napus
- PMID: 33704431
- PMCID: PMC8022990
- DOI: 10.1093/g3journal/jkaa011
Chromosome inheritance and meiotic stability in allopolyploid Brassica napus
Abstract
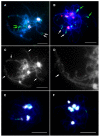
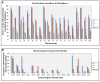
Homoeologous recombination, aneuploidy, and other genetic changes are common in resynthesized allopolyploid Brassica napus. In contrast, the chromosomes of cultivars have long been considered to be meiotically stable. To gain a better understanding of the underlying mechanisms leading to stabilization in the allopolyploid, the behavior of chromosomes during meiosis can be compared by unambiguous chromosome identification between resynthesized and natural B. napus. Compared with natural B. napus, resynthesized lines show high rates of nonhomologous centromere association, homoeologous recombination leading to translocation, homoeologous chromosome replacement, and association and breakage of 45S rDNA loci. In both natural and resynthesized B. napus, we observed low rates of univalents, A-C bivalents, and early sister chromatid separations. Reciprocal homoeologous chromosome exchanges and double reductions were photographed for the first time in meiotic telophase I. Meiotic errors were non-uniformly distributed across the genome in resynthesized B. napus, and in particular homoeologs sharing synteny along their entire length exhibited multivalents at diakinesis and polysomic inheritance at telophase I. Natural B. napus appeared to resolve meiotic errors mainly by suppressing homoeologous pairing, resolving nonhomologous centromere associations and 45S rDNA associations before diakinesis, and reducing homoeologous cross-overs.
Keywords: allopolyploid; double reduction; homoeologous recombination; meiosis error; polysomic inheritance.
© The Author(s) 2020. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
-
- Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB. et al. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 345:950–953. - PubMed
-
- Church K, Moens PB.. 1976. Centromere behavior during interphase and meiotic prophase in Allium fistulosum from 3-D, E.M. reconstruction. Chromosoma. 56:249–263.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
