Regional brain iron and gene expression provide insights into neurodegeneration in Parkinson's disease
- PMID: 33704443
- PMCID: PMC8320305
- DOI: 10.1093/brain/awab084
Regional brain iron and gene expression provide insights into neurodegeneration in Parkinson's disease
Abstract
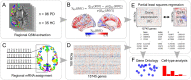
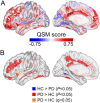
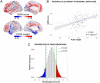
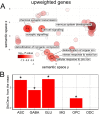
The mechanisms responsible for the selective vulnerability of specific neuronal populations in Parkinson's disease are poorly understood. Oxidative stress secondary to brain iron accumulation is one postulated mechanism. We measured iron deposition in 180 cortical regions of 96 patients with Parkinson's disease and 35 control subjects using quantitative susceptibility mapping. We estimated the expression of 15 745 genes in the same regions using transcriptomic data from the Allen Human Brain Atlas. Using partial least squares regression, we then identified the profile of gene transcription in the healthy brain that underlies increased cortical iron in patients with Parkinson's disease relative to controls. Applying gene ontological tools, we investigated the biological processes and cell types associated with this transcriptomic profile and identified the sets of genes with spatial expression profiles in control brains that correlated significantly with the spatial pattern of cortical iron deposition in Parkinson's disease. Gene ontological analyses revealed that these genes were enriched for biological processes relating to heavy metal detoxification, synaptic function and nervous system development and were predominantly expressed in astrocytes and glutamatergic neurons. Furthermore, we demonstrated that the genes differentially expressed in Parkinson's disease are associated with the pattern of cortical expression identified in this study. Our findings provide mechanistic insights into regional selective vulnerabilities in Parkinson's disease, particularly the processes involving iron accumulation.
Keywords: Parkinson’s disease; genetics; iron; quantitative susceptibility mapping; transcriptomics.
© The Author(s) (2021). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures
References
-
- Hornykiewicz O. Dopamine miracle: From brain homogenate to dopamine replacement. Mov Disord. 2002;17:501-508. - PubMed
-
- Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M.. α-synuclein in Lewy bodies. Nature. 1997;388:839-840. - PubMed
-
- Jellinger KA. A critical evaluation of current staging of α-synuclein pathology in Lewy body disorders. Biochim Biophys Acta Mol Basis Dis. 2009;1792:730-740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

