HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children
- PMID: 33705359
- PMCID: PMC8121516
- DOI: 10.1172/JCI146614
HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children
Abstract
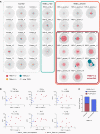
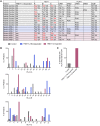
Multisystem inflammatory syndrome in children (MIS-C), a hyperinflammatory syndrome associated with SARS-CoV-2 infection, shares clinical features with toxic shock syndrome, which is triggered by bacterial superantigens. Superantigen specificity for different Vβ chains results in Vβ skewing, whereby T cells with specific Vβ chains and diverse antigen specificity are overrepresented in the T cell receptor (TCR) repertoire. Here, we characterized the TCR repertoire of MIS-C patients and found a profound expansion of TCRβ variable gene 11-2 (TRBV11-2), with up to 24% of clonal T cell space occupied by TRBV11-2 T cells, which correlated with MIS-C severity and serum cytokine levels. Analysis of TRBJ gene usage and complementarity-determining region 3 (CDR3) length distribution of MIS-C expanded TRBV11-2 clones revealed extensive junctional diversity. Patients with TRBV11-2 expansion shared HLA class I alleles A02, B35, and C04, indicating what we believe is a novel mechanism for CDR3-independent T cell expansion. In silico modeling indicated that polyacidic residues in the Vβ chain encoded by TRBV11-2 (Vβ21.3) strongly interact with the superantigen-like motif of SARS-CoV-2 spike glycoprotein, suggesting that unprocessed SARS-CoV-2 spike may directly mediate TRBV11-2 expansion. Overall, our data indicate that a CDR3-independent interaction between SARS-CoV-2 spike and TCR leads to T cell expansion and possibly activation, which may account for the clinical presentation of MIS-C.
Keywords: COVID-19; Immunology; T cell receptor.
Conflict of interest statement
Figures




Comment in
-
SARS-CoV-2 as a superantigen in multisystem inflammatory syndrome in children.J Clin Invest. 2021 May 17;131(10):e149327. doi: 10.1172/JCI149327. J Clin Invest. 2021. PMID: 33844652 Free PMC article.
References
-
- American Academy of Pediatrics. Children and COVID-19: state-level data report. Updated March 8, 2021. Accessed March 9, 2021. https://services.aap.org/en/pages/2019-novel-coronavirus-covid-19-infect...
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

