Microchip RT-PCR Detection of Nasopharyngeal SARS-CoV-2 Samples
- PMID: 33706009
- PMCID: PMC7939975
- DOI: 10.1016/j.jmoldx.2021.02.009
Microchip RT-PCR Detection of Nasopharyngeal SARS-CoV-2 Samples
Abstract
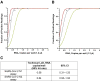
Fast, accurate, and reliable diagnostic tests are critical for controlling the spread of the coronavirus disease 2019 (COVID-19) associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. The current gold standard for testing is real-time PCR; however, during the current pandemic, supplies of testing kits and reagents have been limited. We report the validation of a rapid (30 minutes), user-friendly, and accurate microchip real-time PCR assay for detection of SARS-CoV-2 from nasopharyngeal swab RNA extracts. Microchips preloaded with COVID-19 primers and probes for the N gene accommodate 1.2-μL reaction volumes, lowering the required reagents by 10-fold compared with tube-based real-time PCR. We validated our assay using contrived reference samples and 21 clinical samples from patients in Canada, determining a limit of detection of 1 copy per reaction. The microchip real-time PCR provides a significantly lower resource alternative to the Centers for Disease Control and Prevention-approved real-time RT-PCR assays with comparable sensitivity, showing 100% positive and negative predictive agreement of clinical samples.
Copyright © 2021 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.-R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. - PMC - PubMed
-
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

