Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies
- PMID: 33706364
- PMCID: PMC7616976
- DOI: 10.1038/s41586-021-03412-7
Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies
Erratum in
-
Author Correction: Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies.Nature. 2022 Aug;608(7922):E24. doi: 10.1038/s41586-022-05103-3. Nature. 2022. PMID: 35864232 Free PMC article. No abstract available.
Abstract
Transmission of SARS-CoV-2 is uncontrolled in many parts of the world; control is compounded in some areas by the higher transmission potential of the B.1.1.7 variant1, which has now been reported in 94 countries. It is unclear whether the response of the virus to vaccines against SARS-CoV-2 on the basis of the prototypic strain will be affected by the mutations found in B.1.1.7. Here we assess the immune responses of individuals after vaccination with the mRNA-based vaccine BNT162b22. We measured neutralizing antibody responses after the first and second immunizations using pseudoviruses that expressed the wild-type spike protein or a mutated spike protein that contained the eight amino acid changes found in the B.1.1.7 variant. The sera from individuals who received the vaccine exhibited a broad range of neutralizing titres against the wild-type pseudoviruses that were modestly reduced against the B.1.1.7 variant. This reduction was also evident in sera from some patients who had recovered from COVID-19. Decreased neutralization of the B.1.1.7 variant was also observed for monoclonal antibodies that target the N-terminal domain (9 out of 10) and the receptor-binding motif (5 out of 31), but not for monoclonal antibodies that recognize the receptor-binding domain that bind outside the receptor-binding motif. Introduction of the mutation that encodes the E484K substitution in the B.1.1.7 background to reflect a newly emerged variant of concern (VOC 202102/02) led to a more-substantial loss of neutralizing activity by vaccine-elicited antibodies and monoclonal antibodies (19 out of 31) compared with the loss of neutralizing activity conferred by the mutations in B.1.1.7 alone. The emergence of the E484K substitution in a B.1.1.7 background represents a threat to the efficacy of the BNT162b2 vaccine.
Conflict of interest statement
A.D.M., J.B., D.P., C.S.F., S.B., K.C., N.S., E.C., G.S., S.J., A.L., H.W.V., M.S.P., L.P. and D.C. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. H.W.V. is a founder of PierianDx and Casma Therapeutics. Neither company provided funding for this work or is performing related work. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. RKG has received consulting fees from UMOVIS Lab, Gilead and ViiV.
Figures
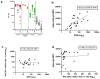

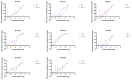



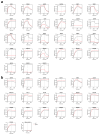





Update of
-
SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies.medRxiv [Preprint]. 2021 Feb 15:2021.01.19.21249840. doi: 10.1101/2021.01.19.21249840. medRxiv. 2021. Update in: Nature. 2021 May;593(7857):136-141. doi: 10.1038/s41586-021-03412-7. PMID: 33619509 Free PMC article. Updated. Preprint.
Comment in
-
What scientists know about new, fast-spreading coronavirus variants.Nature. 2021 Jun;594(7861):19-20. doi: 10.1038/d41586-021-01390-4. Nature. 2021. PMID: 34031583 No abstract available.
References
-
- Davies NG, et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv. 2020:2020.2012.2024.20248822. doi: 10.1101/2020.12.24.20248822. - DOI
-
- Volz E, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021:2020.2012.2030.20249034. doi: 10.1101/2020.12.30.20249034. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- MR/P008801/1/MRC_/Medical Research Council/United Kingdom
- R01 GM120553/GM/NIGMS NIH HHS/United States
- DP1 AI158186/AI/NIAID NIH HHS/United States
- 200871/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- S10 OD023476/OD/NIH HHS/United States
- MR/S00081X/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_19027/MRC_/Medical Research Council/United Kingdom
- MR/R008698/1/MRC_/Medical Research Council/United Kingdom
- 108082/WT_/Wellcome Trust/United Kingdom
- HHSN272201700059C/AI/NIAID NIH HHS/United States
- 204911/Z/16/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

