Trehalose causes low-grade lysosomal stress to activate TFEB and the autophagy-lysosome biogenesis response
- PMID: 33706671
- PMCID: PMC8632292
- DOI: 10.1080/15548627.2021.1896906
Trehalose causes low-grade lysosomal stress to activate TFEB and the autophagy-lysosome biogenesis response
Abstract
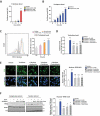
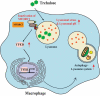
The autophagy-lysosome system is an important cellular degradation pathway that recycles dysfunctional organelles and cytotoxic protein aggregates. A decline in this system is pathogenic in many human diseases including neurodegenerative disorders, fatty liver disease, and atherosclerosis. Thus there is intense interest in discovering therapeutics aimed at stimulating the autophagy-lysosome system. Trehalose is a natural disaccharide composed of two glucose molecules linked by a ɑ-1,1-glycosidic bond with the unique ability to induce cellular macroautophagy/autophagy and with reported efficacy on mitigating several diseases where autophagy is dysfunctional. Interestingly, the mechanism by which trehalose induces autophagy is unknown. One suggested mechanism is its ability to activate TFEB (transcription factor EB), the master transcriptional regulator of autophagy-lysosomal biogenesis. Here we describe a potential mechanism involving direct trehalose action on the lysosome. We find trehalose is endocytically taken up by cells and accumulates within the endolysosomal system. This leads to a low-grade lysosomal stress with mild elevation of lysosomal pH, which acts as a potent stimulus for TFEB activation and nuclear translocation. This process appears to involve inactivation of MTORC1, a known negative regulator of TFEB which is sensitive to perturbations in lysosomal pH. Taken together, our data show the trehalose can act as a weak inhibitor of the lysosome which serves as a trigger for TFEB activation. Our work not only sheds light on trehalose action but suggests that mild alternation of lysosomal pH can be a novel method of inducing the autophagy-lysosome system.Abbreviations: ASO: antisense oligonucleotide; AU: arbitrary units; BMDM: bone marrow-derived macrophages; CLFs: crude lysosomal fractions; CTSD: cathepsin D; LAMP: lysosomal associated membrane protein; LIPA/LAL: lipase A, lysosomal acid type; MAP1LC3: microtubule-associated protein 1 light chain 3; MFI: mean fluorescence intensity; MTORC1: mechanistic target of rapamycin kinase complex 1; pMAC: peritoneal macrophages; SLC2A8/GLUT8: solute carrier family 2, (facilitated glucose transporter), member 8; TFEB: transcription factor EB; TMR: tetramethylrhodamine; TREH: trehalase.
Keywords: Endocytosis; MTORC1; TFEB; lysosome; trehalose.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
-
- Nakatogawa H, Suzuki K, Kamada Y, et al. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol [Internet]. 2009;10:458–467. - PubMed
-
- Mizushima N, Komatsu M.. Review autophagy : renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
-
- Sa´nchez I, Mahlke C, Yuan J.. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. - PubMed
-
- Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. - PubMed
-
- Polanco JC, Li C, Bodea L, et al. Amyloid‑β and tau complexity — towards improved biomarkers and targeted therapies. Nat Rev Neurol. 2018;14:22–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL125838/HL/NHLBI NIH HHS/United States
- R01 DK115867/DK/NIDDK NIH HHS/United States
- T32 HL134635/HL/NHLBI NIH HHS/United States
- R01 DK126622/DK/NIDDK NIH HHS/United States
- R01 HL107594/HL/NHLBI NIH HHS/United States
- P41 GM103422/GM/NIGMS NIH HHS/United States
- R01 HL147968/HL/NHLBI NIH HHS/United States
- P30 DK020579/DK/NIDDK NIH HHS/United States
- I01 BX004235/BX/BLRD VA/United States
- R01 DK118333/DK/NIDDK NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- F31 HL132434/HL/NHLBI NIH HHS/United States
- R21 AT010520/AT/NCCIH NIH HHS/United States
- T32 HL007081/HL/NHLBI NIH HHS/United States
- I01 BX003415/BX/BLRD VA/United States
- R56 DK115764/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
