Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19
- PMID: 33707363
- PMCID: PMC7904510
- DOI: 10.1007/s12041-021-01262-w
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19
Abstract
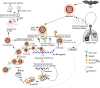
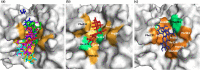
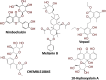
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection is at present an emerging global public health crisis. Angiotensin converting enzyme 2 (ACE2) and trans-membrane protease serine 2 (TMPRSS2) are the two major host factors that contribute to the virulence of SARS-CoV-2 and pathogenesis of coronavirus disease-19 (COVID-19). Transmission of SARS-CoV-2 from animal to human is considered a rare event that necessarily requires strong evolutionary adaptations. Till date no other human cellular receptors are identified beside ACE2 for SARS-CoV-2 entry inside the human cell. Proteolytic cleavage of viral spike (S)-protein and ACE2 by TMPRSS2 began the entire host-pathogen interaction initiated with the physical binding of ACE2 to S-protein. SARS-CoV-2 S-protein binds to ACE2 with much higher affinity and stability than that of SARS-CoVs. Molecular interactions between ACE2-S and TMPRSS2-S are crucial and preciously mediated by specific residues. Structural stability, binding affinity and level of expression of these three interacting proteins are key susceptibility factors for COVID-19. Specific protein-protein interactions (PPI) are being identified that explains uniqueness of SARS-CoV-2 infection. Amino acid substitutions due to naturally occurring genetic polymorphisms potentially alter these PPIs and poses further clinical heterogeneity of COVID-19. Repurposing of several phytochemicals and approved drugs against ACE2, TMPRSS2 and S-protein have been proposed that could inhibit PPI between them. We have also identified some novel lead phytochemicals present in Azadirachta indica and Aloe barbadensis which could be utilized for further in vitro and in vivo anti-COVID-19 drug discovery. Uncovering details of ACE2-S and TMPRSS2-S interactions would further contribute to future research on COVID-19.
Figures


References
-
- Abdel-Mottaleb MS, Abdel-Mottaleb Y. In search for effective and safe drugs against SARS-CoV-2: Part I] simulated interactions between selected nutraceuticals ACE2 enzyme and S protein simple peptide sequences. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12155235.v1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
