SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface
- PMID: 33707427
- PMCID: PMC7952403
- DOI: 10.1038/s41467-021-21609-2
SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface
Abstract
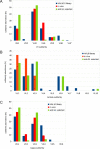
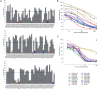
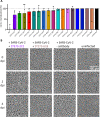
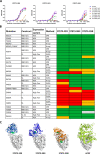
COVID-19 is a severe acute respiratory disease caused by SARS-CoV-2, a new recently emerged sarbecovirus. This virus uses the human ACE2 enzyme as receptor for cell entry, recognizing it with the receptor binding domain (RBD) of the S1 subunit of the viral spike protein. We present the use of phage display to select anti-SARS-CoV-2 spike antibodies from the human naïve antibody gene libraries HAL9/10 and subsequent identification of 309 unique fully human antibodies against S1. 17 antibodies are binding to the RBD, showing inhibition of spike binding to cells expressing ACE2 as scFv-Fc and neutralize active SARS-CoV-2 virus infection of VeroE6 cells. The antibody STE73-2E9 is showing neutralization of active SARS-CoV-2 as IgG and is binding to the ACE2-RBD interface. Thus, universal libraries from healthy human donors offer the advantage that antibodies can be generated quickly and independent from the availability of material from recovering patients in a pandemic situation.
Conflict of interest statement
The authors declare a conflict of interest. The authors F.B., D.M., N.L., S.S., P.A.H., R.B., M.R., K.T.S., K.D.R.R., S.Z.-E., M.B., V.F., S.T., M.S. and M.H. submitted a patent application on blocking antibodies against SARS-CoV-2. The patent was licensed to CORAT Therapeutics GmbH, a company founded by YUMAB GmbH. S.D. and M.H. are shareholders of YUMAB GmbH.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

