Sanger sequencing is no longer always necessary based on a single-center validation of 1109 NGS variants in 825 clinical exomes
- PMID: 33707547
- PMCID: PMC7952542
- DOI: 10.1038/s41598-021-85182-w
Sanger sequencing is no longer always necessary based on a single-center validation of 1109 NGS variants in 825 clinical exomes
Abstract
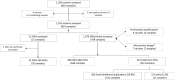
Despite the improved accuracy of next-generation sequencing (NGS), it is widely accepted that variants need to be validated using Sanger sequencing before reporting. Validation of all NGS variants considerably increases the turnaround time and costs of clinical diagnosis. We comprehensively assessed this need in 1109 variants from 825 clinical exomes, the largest sample set to date assessed using Illumina chemistry reported. With a concordance of 100%, we conclude that Sanger sequencing can be very useful as an internal quality control, but not so much as a verification method for high-quality single-nucleotide and small insertion/deletions variants. Laboratories might validate and establish their own thresholds before discontinuing Sanger confirmation studies. We also expand and validate 23 copy number variations detected by exome sequencing in 20 samples, observing a concordance of 95.65% (22/23).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Nelson AC, et al. Criteria for clinical reporting of variants from a broad target capture NGS assay without Sanger verification. JSM Biomark. 2015;2:1005.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources