Plasma metabolites to profile pathways in noncommunicable disease multimorbidity
- PMID: 33707775
- PMCID: PMC8127079
- DOI: 10.1038/s41591-021-01266-0
Plasma metabolites to profile pathways in noncommunicable disease multimorbidity
Abstract
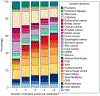
Multimorbidity, the simultaneous presence of multiple chronic conditions, is an increasing global health problem and research into its determinants is of high priority. We used baseline untargeted plasma metabolomics profiling covering >1,000 metabolites as a comprehensive readout of human physiology to characterize pathways associated with and across 27 incident noncommunicable diseases (NCDs) assessed using electronic health record hospitalization and cancer registry data from over 11,000 participants (219,415 person years). We identified 420 metabolites shared between at least 2 NCDs, representing 65.5% of all 640 significant metabolite-disease associations. We integrated baseline data on over 50 diverse clinical risk factors and characteristics to identify actionable shared pathways represented by those metabolites. Our study highlights liver and kidney function, lipid and glucose metabolism, low-grade inflammation, surrogates of gut microbial diversity and specific health-related behaviors as antecedents of common NCD multimorbidity with potential for early prevention. We integrated results into an open-access webserver ( https://omicscience.org/apps/mwasdisease/ ) to facilitate future research and meta-analyses.
Conflict of interest statement
Competing interests
G.A.M. is an employee of Metabolon. All other authors declare no competing interests.
Figures



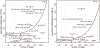








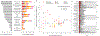


References
Publication types
MeSH terms
Grants and funding
- R01 AG046171/AG/NIA NIH HHS/United States
- MC-UU_12015/1/MRC_/Medical Research Council/United Kingdom
- R01 MH108348/MH/NIMH NIH HHS/United States
- G1000143/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/1/MRC_/Medical Research Council/United Kingdom
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- 14136/CRUK_/Cancer Research UK/United Kingdom
- RF1 AG059093/AG/NIA NIH HHS/United States
- U01 AG061359/AG/NIA NIH HHS/United States
- RF1 AG057452/AG/NIA NIH HHS/United States
- MC_UU_00006/1/MRC_/Medical Research Council/United Kingdom
- U19 AG063744/AG/NIA NIH HHS/United States
- C864/A14136/CRUK_/Cancer Research UK/United Kingdom
- G0401527/MRC_/Medical Research Council/United Kingdom
- MR/L00002/1/MRC_/Medical Research Council/United Kingdom
- RF1 AG058942/AG/NIA NIH HHS/United States
- RF1 AG051550/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

