Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains
- PMID: 33707782
- PMCID: PMC7610442
- DOI: 10.1038/s41589-021-00753-2
Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains
Abstract
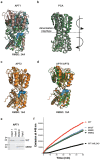
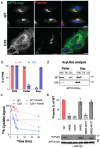
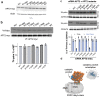
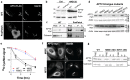
Many biochemical reactions require controlled recruitment of proteins to membranes. This is largely regulated by posttranslational modifications. A frequent one is S-acylation, which consists of the addition of acyl chains and can be reversed by poorly understood acyl protein thioesterases (APTs). Using a panel of computational and experimental approaches, we dissect the mode of action of the major cellular thioesterase APT2 (LYPLA2). We show that soluble APT2 is vulnerable to proteasomal degradation, from which membrane binding protects it. Interaction with membranes requires three consecutive steps: electrostatic attraction, insertion of a hydrophobic loop and S-acylation by the palmitoyltransferases ZDHHC3 or ZDHHC7. Once bound, APT2 is predicted to deform the lipid bilayer to extract the acyl chain bound to its substrate and capture it in a hydrophobic pocket to allow hydrolysis. This molecular understanding of APT2 paves the way to understand the dynamics of APT2-mediated deacylation of substrates throughout the endomembrane system.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Deform the membrane, cAPTure the lipid.Nat Chem Biol. 2021 Apr;17(4):371-372. doi: 10.1038/s41589-021-00754-1. Nat Chem Biol. 2021. PMID: 33707783 No abstract available.
References
-
- Blanc M, David FPA, van der Goot FG. SwissPalm 2: Protein S-Palmitoylation Database. Methods Mol Biol. 2019;2009:203–214. - PubMed
-
- Gadalla MR, Veit M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opinion on Drug Discovery. 2020;15:159–177. - PubMed
-
- Zaballa M-E, van der Goot FG. The molecular era of protein S-acylation: spotlight on structure, mechanisms, and dynamics. Crit Rev Biochem Mol Biol. 2018;53:420–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

