Longitudinal Study of the Bulk Tank Milk Microbiota Reveals Major Temporal Shifts in Composition
- PMID: 33708181
- PMCID: PMC7940241
- DOI: 10.3389/fmicb.2021.616429
Longitudinal Study of the Bulk Tank Milk Microbiota Reveals Major Temporal Shifts in Composition
Abstract
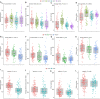
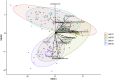
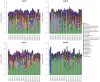
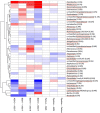
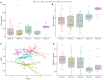
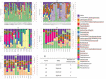
Introduction of microbial contaminations in the dairy value chain starts at the farm level and the initial microbial composition may severely affect the production of high-quality dairy products. Therefore, understanding the farm-to-farm variation and longitudinal shifts in the composition of the bulk tank milk microbiota is fundamental to increase the quality and reduce the spoilage and waste of milk and dairy products. In this study, we performed a double experiment to study long- and short-term longitudinal shifts in microbial composition using 16S rRNA gene amplicon sequencing. We analyzed milk from 37 farms, that had also been investigated two years earlier, to understand the stability and overall microbial changes over a longer time span. In addition, we sampled bulk tank milk from five farms every 1-2 weeks for up to 7 months to observe short-term changes in microbial composition. We demonstrated that a persistent and farm-specific microbiota is found in bulk tank milk and that changes in composition within the same farm are mostly driven by bacterial genera associated with mastitis (e.g., Staphylococcus and Streptococcus). On a long-term, we detected that major shift in milk microbiota were not correlated with farm settings, such as milking system, number of cows and quality of the milk but other factors, such as weather and feeding, may have had a greater impact on the main shifts in composition of the bulk tank milk microbiota. Our results provide new information regarding the ecology of raw milk microbiota at the farm level.
Keywords: bulk tank; longitudinal study; milk quality; raw milk microbiota; sequence variants.
Copyright © 2021 Porcellato, Smistad, Bombelli, Abdelghani, Jørgensen and Skeie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Falentin H., Rault L., Nicolas A., Bouchard D. S., Lassalas J., Lamberton P., et al. (2016). Bovine teat microbiome analysis revealed reduced alpha diversity and significant changes in taxonomic profiles in quarters with a history of mastitis. Front. Microbiol 7:480. 10.3389/fmicb.2016.00480 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

