VJ Segment Usage of TCR-Beta Repertoire in Monozygotic Cystic Fibrosis Twins
- PMID: 33708199
- PMCID: PMC7940196
- DOI: 10.3389/fimmu.2021.599133
VJ Segment Usage of TCR-Beta Repertoire in Monozygotic Cystic Fibrosis Twins
Abstract
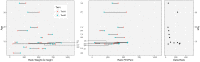
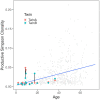
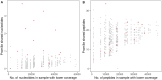
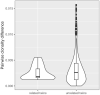
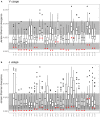
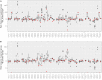
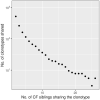
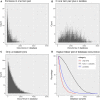
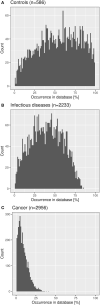
Sixteen monozygotic cystic fibrosis (CF) twin pairs of whom 14 pairs were homozygous for the most common p.Phe508del CFTR mutation were selected from the European Cystic Fibrosis Twin and Sibling Study Cohort. The monozygotic twins were examined in their T cell receptor (TCR) repertoire in peripheral blood by amplicon sequencing of the CDR3 variable region of the ß-chain. The recruitment of TCR J and V genes for recombination and selection in the thymus showed a strong genetic influence in the CF twin cohort as indicated by the shortest Jensen-Shannon distance to the twin individual. Exceptions were the clinically most discordant and/or most severely affected twin pairs where clonal expansion probably caused by recurrent pulmonary infections overshadowed the impact of the identical genomic blueprint. In general the Simpson clonality was low indicating that the population of TCRß clonotypes of the CF twins was dominated by the naïve T-cell repertoire. Intrapair sharing of clonotypes was significantly more frequent among monozygotic CF twins than among pairs of unrelated CF patients. Complete nucleotide sequence identity was observed in about 0.11% of CDR3 sequences which partially should represent persisting fetal clones derived from the same progenitor T cells. Complete amino acid sequence identity was noted in 0.59% of clonotypes. Of the nearly 40,000 frequent amino acid clonotypes shared by at least two twin siblings 99.8% were already known within the immuneACCESS database and only 73 had yet not been detected indicating that the CDR3ß repertoire of CF children and adolescents does not carry a disease-specific signature but rather shares public clones with that of the non-CF community. Clonotypes shared within twin pairs and between unrelated CF siblings were highly abundant among healthy non-CF people, less represented in individuals with infectious disease and uncommon in patients with cancer. This subset of shared CF clonotypes defines CDR3 amino acid sequences that are more common in health than in disease.
Keywords: CDR3; T cell receptor repertoire; TCRB; VJ usage; cyctic fibrosis; immunotyping; twins.
Copyright © 2021 Fischer, Stanke and Tümmler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Determinants governing T cell receptor α/β-chain pairing in repertoire formation of identical twins.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):532-540. doi: 10.1073/pnas.1915008117. Epub 2019 Dec 26. Proc Natl Acad Sci U S A. 2020. PMID: 31879353 Free PMC article.
-
Immunotyping of clinically divergent p.Phe508del homozygous monozygous cystic fibrosis twins.J Cyst Fibros. 2021 Jan;20(1):149-153. doi: 10.1016/j.jcf.2020.06.009. Epub 2020 Jun 13. J Cyst Fibros. 2021. PMID: 32540173
-
Characterization of CD8+ T cell repertoire in identical twins discordant and concordant for multiple sclerosis.J Leukoc Biol. 2007 Mar;81(3):696-710. doi: 10.1189/jlb.0906584. Epub 2006 Nov 16. J Leukoc Biol. 2007. PMID: 17110420
-
Application of the molecular analysis of the T-cell receptor repertoire in the study of immune-mediated hematologic diseases.Hematology. 2003 Jun;8(3):173-81. doi: 10.1080/1024533031000107505. Hematology. 2003. PMID: 12745651 Review.
-
The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor.Front Immunol. 2013 Sep 2;4:263. doi: 10.3389/fimmu.2013.00263. Front Immunol. 2013. PMID: 24032032 Free PMC article. Review.
Cited by
-
TCR repertoire dynamics and their responses underscores dengue severity.iScience. 2024 Sep 16;27(10):110983. doi: 10.1016/j.isci.2024.110983. eCollection 2024 Oct 18. iScience. 2024. PMID: 39403197 Free PMC article.
-
Quantifiable blood TCR repertoire components associate with immune aging.Nat Commun. 2024 Sep 17;15(1):8171. doi: 10.1038/s41467-024-52522-z. Nat Commun. 2024. PMID: 39289351 Free PMC article.
-
Combining genotypes and T cell receptor distributions to infer genetic loci determining V(D)J recombination probabilities.Elife. 2022 Mar 22;11:e73475. doi: 10.7554/eLife.73475. Elife. 2022. PMID: 35315770 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

