Bistable Mathematical Model of Neutrophil Migratory Patterns After LPS-Induced Epigenetic Reprogramming
- PMID: 33708241
- PMCID: PMC7940759
- DOI: 10.3389/fgene.2021.633963
Bistable Mathematical Model of Neutrophil Migratory Patterns After LPS-Induced Epigenetic Reprogramming
Abstract
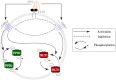
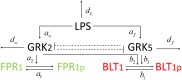
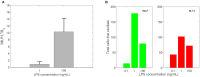
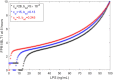
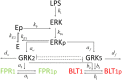
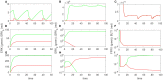
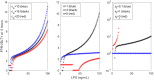
The highly controlled migration of neutrophils toward the site of an infection can be altered when they are trained with lipopolysaccharides (LPS), with high dose LPS enhancing neutrophil migratory pattern toward the bacterial derived source signal and super-low dose LPS inducing either migration toward an intermediary signal or dysregulation and oscillatory movement. Empirical studies that use microfluidic chemotaxis-chip devices with two opposing chemoattractants showed differential neutrophil migration after challenge with different LPS doses. The epigenetic alterations responsible for changes in neutrophil migratory behavior are unknown. We developed two mathematical models that evaluate the mechanistic interactions responsible for neutrophil migratory decision-making when exposed to competing chemoattractants and challenged with LPS. The first model, which considers the interactions between the receptor densities of two competing chemoattractants, their kinases, and LPS, displayed bistability between high and low ratios of primary to intermediary chemoattractant receptor densities. In particular, at equilibrium, we observe equal receptor densities for low LPS (< 15ng/mL); and dominance of receptors for the primary chemoattractant for high LPS (> 15ng/mL). The second model, which included additional interactions with an extracellular signal-regulated kinase in both phosphorylated and non-phosphorylated forms, has an additional dynamic outcome, oscillatory dynamics for both receptors, as seen in the data. In particular, it found equal receptor densities in the absence of oscillation for super-low and high LPS challenge (< 0.4 and 1.1 <LPS< 375 ng/mL); equal receptor densities with oscillatory receptor dynamics for super-low LPS (0.5 < LPS< 1.1ng/mL); and dominance of receptors for the primary chemoattractant for super-high LPS (>376 ng/mL). Predicting the mechanisms and the type of external LPS challenge responsible for neutrophils migration toward pro-inflammatory chemoattractants, migration toward pro-tolerant chemoattractants, or oscillatory movement is necessary knowledge in designing interventions against immune diseases, such as sepsis.
Keywords: bistability; cellular decision-making; lipopolysaccharide (LPS); mathematical model; neutrophil migration.
Copyright © 2021 Ciupe, Boribong, Kadelka and Jones.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Super-Low Dose Lipopolysaccharide Dysregulates Neutrophil Migratory Decision-Making.Front Immunol. 2019 Mar 12;10:359. doi: 10.3389/fimmu.2019.00359. eCollection 2019. Front Immunol. 2019. PMID: 30915068 Free PMC article.
-
An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients.J Cell Biol. 2002 Oct 14;159(1):91-102. doi: 10.1083/jcb.200202114. Epub 2002 Oct 7. J Cell Biol. 2002. PMID: 12370241 Free PMC article.
-
Endotoxin promotes neutrophil hierarchical chemotaxis via the p38-membrane receptor pathway.Oncotarget. 2016 Nov 8;7(45):74247-74258. doi: 10.18632/oncotarget.12093. Oncotarget. 2016. PMID: 27655676 Free PMC article.
-
The function of neutrophils in sepsis.Curr Opin Infect Dis. 2012 Jun;25(3):321-7. doi: 10.1097/QCO.0b013e3283528c9b. Curr Opin Infect Dis. 2012. PMID: 22421753 Review.
-
Neutrophil chemotaxis.Cell Tissue Res. 2018 Mar;371(3):425-436. doi: 10.1007/s00441-017-2776-8. Epub 2018 Jan 19. Cell Tissue Res. 2018. PMID: 29350282 Review.
Cited by
-
The dual role of neutrophils in sepsis-associated liver injury.Front Immunol. 2025 Feb 28;16:1538282. doi: 10.3389/fimmu.2025.1538282. eCollection 2025. Front Immunol. 2025. PMID: 40092997 Free PMC article. Review.
References
-
- Arraes S. M. A., Freitas M. S., da Silva S. V., de Paula Neto H. A., Alves-Filho J. C., Martins M. A., et al. . (2006). Impaired neutrophil chemotaxis in sepsis associates with grk expression and inhibition of actin assembly and tyrosine phosphorylation. Blood 108, 2906–2913. 10.1182/blood-2006-05-024638 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

