Circular RNA TRAPPC6B inhibits intracellular Mycobacterium tuberculosis growth while inducing autophagy in macrophages by targeting microRNA-874-3p
- PMID: 33708385
- PMCID: PMC7890665
- DOI: 10.1002/cti2.1254
Circular RNA TRAPPC6B inhibits intracellular Mycobacterium tuberculosis growth while inducing autophagy in macrophages by targeting microRNA-874-3p
Abstract
Objectives: Genetic and epigenetic mechanisms regulate antimicrobial immunity against Mycobacterium tuberculosis (Mtb) infection.
Methods: The present study assessed circular RNA TRAPPC6B (circTRAPPC6B) for antimicrobial immune functions and defined mechanisms wherein circTRAPPC6B regulates Mtb growth, autophagy and microRNA in macrophages.
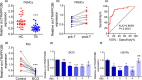
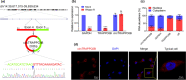
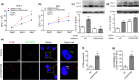
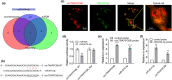
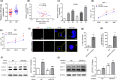
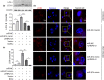
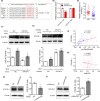
Results: The Mtb infection of monocytes/macrophages resulted in a significantly decreased level of circTRAPPC6B that inhibited intracellular Mtb growth in macrophages. Conversely, circTRAPPC6B expression enhanced autophagy or autophagy-associated protein LC3-II production in Mtb-infected macrophages. circTRAPPC6B-enhanced autophagy aggregation or sequestration was also observed in fluorescence in situ hybridisation (FISH) analysis and confocal imaging. Mechanistically, circTRAPPC6B targets an inhibiting element miR-874-3p, as shown by bioinformatics, dual-luciferase reporter gene analysis and pull-down assay, respectively. Notably, miR-874-3p prohibited autophagy via suppressing autophagy protein ATG16L1 by binding to its 3'-untranslated region (UTR) in Mtb-infected macrophages and thus promoting intracellular Mtb growth. Concurrently, circTRAPPC6B enhanced autophagy in Mtb-infected macrophages by blocking the ability of miR-874-3p to inhibit ATG16L1. Thus, circTRAPPC6B antagonises the ability of miR-874-3p to suppress ATG16L1 expression and activate and enhance autophagy sequestration to restrict Mtb growth in macrophages.
Conclusion: The current findings suggested that both circTRAPPC6B and miR-874-3p mechanisms can be explored as potential therapeutics against Mtb infection.
Keywords: Mycobacterium tuberculosis; autophagy; circTRAPPC6B; macrophage; miR‐874‐3p.
© 2021 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
