TBL1XR1 Ensures Balanced Neural Development Through NCOR Complex-Mediated Regulation of the MAPK Pathway
- PMID: 33708771
- PMCID: PMC7940385
- DOI: 10.3389/fcell.2021.641410
TBL1XR1 Ensures Balanced Neural Development Through NCOR Complex-Mediated Regulation of the MAPK Pathway
Abstract
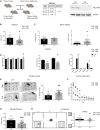
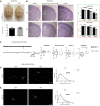
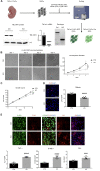
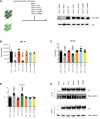
TBL1XR1 gene is associated with multiple developmental disorders presenting several neurological aspects. The relative protein is involved in the modulation of important cellular pathways and master regulators of transcriptional output, including nuclear receptor repressors, Wnt signaling, and MECP2 protein. However, TBL1XR1 mutations (including complete loss of its functions) have not been experimentally studied in a neurological context, leaving a knowledge gap in the mechanisms at the basis of the diseases. Here, we show that Tbl1xr1 knock-out mice exhibit behavioral and neuronal abnormalities. Either the absence of TBL1XR1 or its point mutations interfering with stability/regulation of NCOR complex induced decreased proliferation and increased differentiation in neural progenitors. We suggest that this developmental unbalance is due to a failure in the regulation of the MAPK cascade. Taken together, our results broaden the molecular and functional aftermath of TBL1XR1 deficiency associated with human disorders.
Keywords: MAPK; NCOR; TBL1XR1; brain development; neurodevelopmental disorders.
Copyright © 2021 Mastrototaro, Zaghi, Massimino, Moneta, Mohammadi, Banfi, Bellini, Indrigo, Fagnocchi, Bagliani, Taverna, Rohm, Herzig and Sessa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
TBL1XR1 in physiological and pathological states.Am J Clin Exp Urol. 2015 Apr 25;3(1):13-23. eCollection 2015. Am J Clin Exp Urol. 2015. PMID: 26069883 Free PMC article. Review.
-
Analysis of genes differentially expressed in the cortex of mice with the Tbl1xr1Y446C/Y446C variant.Gene. 2024 Nov 15;927:148707. doi: 10.1016/j.gene.2024.148707. Epub 2024 Jun 15. Gene. 2024. PMID: 38885822
-
A case of West syndrome and global developmental delay in a child with a heterozygous mutation in the TBL1XR1 gene: A case report.Medicine (Baltimore). 2023 May 12;102(19):e33744. doi: 10.1097/MD.0000000000033744. Medicine (Baltimore). 2023. PMID: 37171308 Free PMC article.
-
The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP.Oncotarget. 2017 Mar 7;8(10):17038-17049. doi: 10.18632/oncotarget.14928. Oncotarget. 2017. PMID: 28152507 Free PMC article.
-
Two decades of a protooncogene TBL1XR1: from a transcription modulator to cancer therapeutic target.Front Oncol. 2024 Jan 29;14:1309687. doi: 10.3389/fonc.2024.1309687. eCollection 2024. Front Oncol. 2024. PMID: 38347836 Free PMC article. Review.
Cited by
-
Autism risk genes converge on PBX1 to govern neural cell growth.bioRxiv [Preprint]. 2025 Mar 13:2025.03.12.642693. doi: 10.1101/2025.03.12.642693. bioRxiv. 2025. PMID: 40161581 Free PMC article. Preprint.
-
Deregulation of ceRNA Networks in Frontal Cortex and Choroid Plexus of Brain during SARS-CoV-2 Infection Aggravates Neurological Manifestations: An Insight from Bulk and Single-Cell Transcriptomic Analyses.Adv Biol (Weinh). 2022 Aug;6(8):e2101310. doi: 10.1002/adbi.202101310. Epub 2022 Jun 3. Adv Biol (Weinh). 2022. PMID: 35661455 Free PMC article.
-
Investigating milk-derived extracellular vesicles as mediators of maternal stress and environmental intervention.bioRxiv [Preprint]. 2025 Jun 27:2025.05.30.656911. doi: 10.1101/2025.05.30.656911. bioRxiv. 2025. PMID: 40502175 Free PMC article. Preprint.
-
Excess Wnt in neurological disease.Biochem J. 2025 May 16;482(10):601-18. doi: 10.1042/BCJ20240265. Biochem J. 2025. PMID: 40377402 Free PMC article. Review.
-
Population structure and selection signal analysis of indigenous sheep from the southern edge of the Taklamakan Desert.BMC Genomics. 2024 Jul 9;25(1):681. doi: 10.1186/s12864-024-10581-y. BMC Genomics. 2024. PMID: 38982349 Free PMC article.
References
-
- Castelo-Branco G., Lilja T., Wallenborg K., Falcao A. M., Marques S. C., Gracias A., et al. (2014). Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Reports 3 502–515. 10.1016/j.stemcr.2014.07.008 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

