Polygenic Risk Scores Augment Stroke Subtyping
- PMID: 33709033
- PMCID: PMC7943221
- DOI: 10.1212/NXG.0000000000000560
Polygenic Risk Scores Augment Stroke Subtyping
Abstract
Objective: To determine whether the polygenic risk score (PRS) derived from MEGASTROKE is associated with ischemic stroke (IS) and its subtypes in an independent tertiary health care system and to identify the PRS derived from gene sets of known biological pathways associated with IS.
Methods: Controls (n = 19,806/7,484, age ≥69/79 years) and cases (n = 1,184/951 for discovery/replication) of acute IS with European ancestry and clinical risk factors were identified by leveraging the Geisinger Electronic Health Record and chart review confirmation. All Geisinger MyCode patients with age ≥69/79 years and without any stroke-related diagnostic codes were included as low risk control. Genetic heritability and genetic correlation between Geisinger and MEGASTROKE (EUR) were calculated using the summary statistics of the genome-wide association study by linkage disequilibrium score regression. All PRS for any stroke (AS), any ischemic stroke (AIS), large artery stroke (LAS), cardioembolic stroke (CES), and small vessel stroke (SVS) were constructed by PRSice-2.
Results: A moderate heritability (10%-20%) for Geisinger sample as well as the genetic correlation between MEGASTROKE and the Geisinger cohort was identified. Variation of all 5 PRS significantly explained some of the phenotypic variations of Geisinger IS, and the R 2 increased by raising the cutoff for the age of controls. PRSLAS, PRSCES, and PRSSVS derived from low-frequency common variants provided the best fit for modeling (R 2 = 0.015 for PRSLAS). Gene sets analyses highlighted the association of PRS with Gene Ontology terms (vascular endothelial growth factor, amyloid precursor protein, and atherosclerosis). The PRSLAS, PRSCES, and PRSSVS explained the most variance of the corresponding subtypes of Geisinger IS suggesting shared etiologies and corroborated Geisinger TOAST subtyping.
Conclusions: We provide the first evidence that PRSs derived from MEGASTROKE have value in identifying shared etiologies and determining stroke subtypes.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Figures
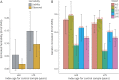
 , A) and genetic correlation (rg, B) were calculated by LDSC using genotyped HapMap 3 SNPs (hm3). Both the observed scale and the liability scale h2, later of which was adjusted by the sample prevalence and population prevalence, were presented in the y-axis, including error bars for the estimates. We assumed a trait prevalence of 1% for all phenotypes and tested the robustness of heritability (
, A) and genetic correlation (rg, B) were calculated by LDSC using genotyped HapMap 3 SNPs (hm3). Both the observed scale and the liability scale h2, later of which was adjusted by the sample prevalence and population prevalence, were presented in the y-axis, including error bars for the estimates. We assumed a trait prevalence of 1% for all phenotypes and tested the robustness of heritability ( ) under 2 levels of controls. AIS = any ischemic stroke; AS = any stroke; CES = cardioembolic stroke; LAS = large artery stroke; LDSC = linkage disequilibrium score regression; SNP = single nucleotide polymorphism; SVS = small vessel stroke.
) under 2 levels of controls. AIS = any ischemic stroke; AS = any stroke; CES = cardioembolic stroke; LAS = large artery stroke; LDSC = linkage disequilibrium score regression; SNP = single nucleotide polymorphism; SVS = small vessel stroke.


References
-
- Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, the Stroke Genetics Network (SiGN), and the International Stroke Genetics Consortium (ISGC). Identification of additional risk loci for stroke and small vessel disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2016;15:695–707. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
