A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure
- PMID: 33710875
- PMCID: PMC8791450
- DOI: 10.1021/acsinfecdis.0c00437
A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure
Abstract
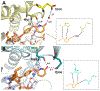
The enoyl-acyl carrier protein (ACP) reductase (ENR) is a key enzyme within the bacterial fatty-acid synthesis pathway. It has been demonstrated that small-molecule inhibitors carrying the diphenylether (DPE) scaffold bear a great potential for the development of highly specific and effective drugs against this enzyme class. Interestingly, different substitution patterns of the DPE scaffold have been shown to lead to varying effects on the kinetic and thermodynamic behavior toward ENRs from different organisms. Here, we investigated the effect of a 4'-pyridone substituent in the context of the slow tight-binding inhibitor SKTS1 on the inhibition of the Staphylococcus aureus enoyl-ACP-reductase saFabI and the closely related isoenzyme from Mycobacterium tuberculosis, InhA, and explored a new interaction site of DPE inhibitors within the substrate-binding pocket. Using high-resolution crystal structures of both complexes in combination with molecular dynamics (MD) simulations, kinetic measurements, and quantum mechanical (QM) calculations, we provide evidence that the 4'-pyridone substituent adopts different tautomeric forms when bound to the two ENRs. We furthermore elucidate the structural determinants leading to significant differences in the residence time of SKTS1 on both enzymes.
Keywords: Mycobacterium tuberculosis; Staphylococcus aureus; diphenylether; enoyl-ACP reductase; residence time; tautomerization.
Figures
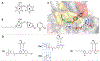
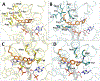



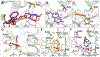
Similar articles
-
A slow, tight binding inhibitor of InhA, the enoyl-acyl carrier protein reductase from Mycobacterium tuberculosis.J Biol Chem. 2010 May 7;285(19):14330-7. doi: 10.1074/jbc.M109.090373. Epub 2010 Mar 3. J Biol Chem. 2010. PMID: 20200152 Free PMC article.
-
A structural and energetic model for the slow-onset inhibition of the Mycobacterium tuberculosis enoyl-ACP reductase InhA.ACS Chem Biol. 2014 Apr 18;9(4):986-93. doi: 10.1021/cb400896g. Epub 2014 Mar 10. ACS Chem Biol. 2014. PMID: 24527857 Free PMC article.
-
Mechanism and inhibition of saFabI, the enoyl reductase from Staphylococcus aureus.Biochemistry. 2008 Apr 8;47(14):4228-36. doi: 10.1021/bi800023a. Epub 2008 Mar 13. Biochemistry. 2008. PMID: 18335995 Free PMC article.
-
Recent Advances and Structural Features of Enoyl-ACP Reductase Inhibitors of Mycobacterium tuberculosis.Arch Pharm (Weinheim). 2016 Nov;349(11):817-826. doi: 10.1002/ardp.201600186. Epub 2016 Oct 24. Arch Pharm (Weinheim). 2016. PMID: 27775177 Review.
-
Targeting InhA, the FASII enoyl-ACP reductase: SAR studies on novel inhibitor scaffolds.Curr Top Med Chem. 2012;12(7):672-93. doi: 10.2174/156802612799984535. Curr Top Med Chem. 2012. PMID: 22283812 Free PMC article. Review.
Cited by
-
Residence time in drug discovery: current insights and future perspectives.Pharmacol Rep. 2025 Aug;77(4):851-873. doi: 10.1007/s43440-025-00748-z. Epub 2025 Jun 9. Pharmacol Rep. 2025. PMID: 40489055 Free PMC article. Review.
-
Is Mycobacterial InhA a Suitable Target for Rational Drug Design?ChemMedChem. 2025 Jul 1;20(13):e202500079. doi: 10.1002/cmdc.202500079. Epub 2025 Apr 29. ChemMedChem. 2025. PMID: 40192582 Free PMC article. Review.
-
Evaluating the Impact of the Tyr158 pKa on the Mechanism and Inhibition of InhA, the Enoyl-ACP Reductase from Mycobacterium tuberculosis.Biochemistry. 2023 Jun 20;62(12):1943-1952. doi: 10.1021/acs.biochem.2c00606. Epub 2023 Jun 4. Biochemistry. 2023. PMID: 37270808 Free PMC article.
References
-
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, and Nouwen JL, The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis, 2005. 5:751–762. - PubMed
-
- King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, and Blumberg HM, Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med, 2006. 144:309–317. - PubMed
-
- Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O’Boyle C, Danila RN, and Lynfield R, Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. Jama, 2003. 290:2976–2984. - PubMed
-
- Pantosti A and Venditti M, What is MRSA? Eur Respir J, 2009. 34:1190–1196. - PubMed
-
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, and Piddock LJ, Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol, 2015. 13:42–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

