A novel cell culture system modeling the SARS-CoV-2 life cycle
- PMID: 33711082
- PMCID: PMC7990224
- DOI: 10.1371/journal.ppat.1009439
A novel cell culture system modeling the SARS-CoV-2 life cycle
Abstract
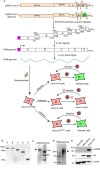
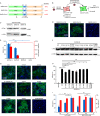
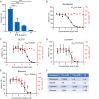
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the global pandemic of COVID-19. SARS-CoV-2 is classified as a biosafety level-3 (BSL-3) agent, impeding the basic research into its biology and the development of effective antivirals. Here, we developed a biosafety level-2 (BSL-2) cell culture system for production of transcription and replication-competent SARS-CoV-2 virus-like-particles (trVLP). This trVLP expresses a reporter gene (GFP) replacing viral nucleocapsid gene (N), which is required for viral genome packaging and virion assembly (SARS-CoV-2 GFP/ΔN trVLP). The complete viral life cycle can be achieved and exclusively confined in the cells ectopically expressing SARS-CoV or SARS-CoV-2 N proteins, but not MERS-CoV N. Genetic recombination of N supplied in trans into viral genome was not detected, as evidenced by sequence analysis after one-month serial passages in the N-expressing cells. Moreover, intein-mediated protein trans-splicing approach was utilized to split the viral N gene into two independent vectors, and the ligated viral N protein could function in trans to recapitulate entire viral life cycle, further securing the biosafety of this cell culture model. Based on this BSL-2 SARS-CoV-2 cell culture model, we developed a 96-well format high throughput screening for antivirals discovery. We identified salinomycin, tubeimoside I, monensin sodium, lycorine chloride and nigericin sodium as potent antivirals against SARS-CoV-2 infection. Collectively, we developed a convenient and efficient SARS-CoV-2 reverse genetics tool to dissect the virus life cycle under a BSL-2 condition. This powerful tool should accelerate our understanding of SARS-CoV-2 biology and its antiviral development.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Q.D. and X.J. have filed a patent application on the use of the SARS-CoV-2 transcomplementation system and its use for anti-SARS-CoV-2 drug screening.
Figures

Comment in
-
Two courses of deconstructed coronavirus please.PLoS Pathog. 2021 Apr 29;17(4):e1009547. doi: 10.1371/journal.ppat.1009547. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33914844 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

