HectD1 controls hematopoietic stem cell regeneration by coordinating ribosome assembly and protein synthesis
- PMID: 33711283
- PMCID: PMC8254759
- DOI: 10.1016/j.stem.2021.02.008
HectD1 controls hematopoietic stem cell regeneration by coordinating ribosome assembly and protein synthesis
Abstract
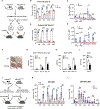
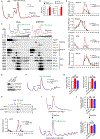
Impaired ribosome function is the underlying etiology in a group of bone marrow failure syndromes called ribosomopathies. However, how ribosomes are regulated remains poorly understood, as are approaches to restore hematopoietic stem cell (HSC) function loss because of defective ribosome biogenesis. Here we reveal a role of the E3 ubiquitin ligase HectD1 in regulating HSC function via ribosome assembly and protein translation. Hectd1-deficient HSCs exhibit a striking defect in transplantation ability and ex vivo maintenance concomitant with reduced protein synthesis and growth rate under stress conditions. Mechanistically, HectD1 ubiquitinates and degrades ZNF622, an assembly factor for the ribosomal 60S subunit. Hectd1 loss leads to accumulation of ZNF622 and the anti-association factor eIF6 on 60S, resulting in 60S/40S joining defects. Importantly, Znf622 depletion in Hectd1-deficient HSCs restored ribosomal subunit joining, protein synthesis, and HSC reconstitution capacity. These findings highlight the importance of ubiquitin-coordinated ribosome assembly in HSC regeneration.
Keywords: HSC regeneration; HectD1; Polypeptide exit tunnel; ZNF622; hematopoietic stem cells; protein synthesis; ribosome assembly; ribosome biogenesis; signaling; ubiquitin.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Releasing the brake on protein synthesis in hematopoietic stem cells.Cell Stem Cell. 2021 Jul 1;28(7):1183-1185. doi: 10.1016/j.stem.2021.06.003. Cell Stem Cell. 2021. PMID: 34214436
References
-
- Aleidi SM, Yang A, Sharpe LJ, Rao G, Cochran BJ, Rye KA, Kockx M, Brown AJ, and Gelissen IC (2018). The E3 ubiquitin ligase, HECTD1, is involved in ABCA1-mediated cholesterol export from macrophages. Biochim Biophys Acta Mol Cell Biol Lipids 1863, 359–368. - PubMed
-
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, and Rommens JM (2003). Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33, 97–101. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

