Strain-based and sex-biased differences in adrenal and pancreatic gene expression between KK/HlJ and C57BL/6 J mice
- PMID: 33711921
- PMCID: PMC7953684
- DOI: 10.1186/s12864-021-07495-4
Strain-based and sex-biased differences in adrenal and pancreatic gene expression between KK/HlJ and C57BL/6 J mice
Abstract
Background: The ever-increasing prevalence of diabetes and associated comorbidities serves to highlight the necessity of biologically relevant small-animal models to investigate its etiology, pathology and treatment. Although the C57BL/6 J model is amongst the most widely used mouse model due to its susceptibility to diet-induced obesity (DIO), there are a number of limitations namely [1] that unambiguous fasting hyperglycemia can only be achieved via dietary manipulation and/or chemical ablation of the pancreatic beta cells. [2] Heterogeneity in the obesogenic effects of hypercaloric feeding has been noted, together with sex-dependent differences, with males being more responsive. The KK mouse strain has been used to study aspects of the metabolic syndrome and prediabetes. We recently conducted a study which characterized the differences in male and female glucocentric parameters between the KK/HlJ and C57BL/6 J strains as well as diabetes-related behavioral differences (Inglis et al. 2019). In the present study, we further characterize these models by examining strain- and sex-dependent differences in pancreatic and adrenal gene expression using Affymetrix microarray together with endocrine-associated serum analysis.
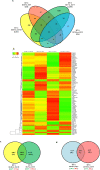
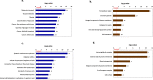
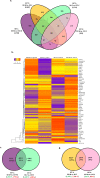
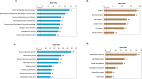
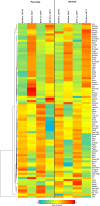
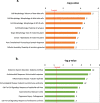
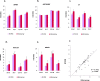
Results: In addition to strain-associated differences in insulin tolerance, we found significant elevations in KK/HlJ mouse serum leptin, insulin and aldosterone. Additionally, glucagon and corticosterone were elevated in female mice of both strains. Using 2-factor ANOVA and a significance level set at 0.05, we identified 10,269 pancreatic and 10,338 adrenal genes with an intensity cut-off of ≥2.0 for all 4 experimental groups. In the pancreas, gene expression upregulated in the KK/HlJ strain related to increased insulin secretory granule biofunction and pancreatic hyperplasia, whereas ontology of upregulated adrenal differentially expressed genes (DEGs) related to cell signaling and neurotransmission. We established a network of functionally related DEGs commonly upregulated in both endocrine tissues of KK/HlJ mice which included the genes coding for endocrine secretory vesicle biogenesis and regulation: PCSK2, PCSK1N, SCG5, PTPRN, CHGB and APLP1. We also identified genes with sex-biased expression common to both strains and tissues including the paternally expressed imprint gene neuronatin.
Conclusion: Our novel results have further characterized the commonalities and diversities of pancreatic and adrenal gene expression between the KK/HlJ and C57BL/6 J strains as well as differences in serum markers of endocrine physiology.
Keywords: C57BL/6 J; Gene expression; Glucose homeostasis; Insulin tolerance test; KK/HlJ; Microarray; Sex-dependent; Strain.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Strain and sex-based glucocentric & behavioral differences between KK/HlJ and C57BL/6J mice.Physiol Behav. 2019 Oct 15;210:112646. doi: 10.1016/j.physbeh.2019.112646. Epub 2019 Aug 7. Physiol Behav. 2019. PMID: 31400379
-
Rhein lysinate decreases inflammation and adipose infiltration in KK/HlJ diabetic mice with non-alcoholic fatty liver disease.Arch Pharm Res. 2016 Jul;39(7):960-9. doi: 10.1007/s12272-016-0770-4. Epub 2016 Jun 8. Arch Pharm Res. 2016. PMID: 27277164
-
Rhein lysinate protects renal function in diabetic nephropathy of KK/HlJ mice.Exp Ther Med. 2017 Dec;14(6):5801-5808. doi: 10.3892/etm.2017.5283. Epub 2017 Oct 11. Exp Ther Med. 2017. PMID: 29285124 Free PMC article.
-
Hereditary diabetes in the KK mouse: an overview.Adv Exp Med Biol. 1988;246:7-15. doi: 10.1007/978-1-4684-5616-5_2. Adv Exp Med Biol. 1988. PMID: 3074669 Review.
-
Knockout mouse models of insulin signaling: Relevance past and future.World J Diabetes. 2014 Apr 15;5(2):146-59. doi: 10.4239/wjd.v5.i2.146. World J Diabetes. 2014. PMID: 24748928 Free PMC article. Review.
Cited by
-
Comprehensive alpha, beta, and delta cell transcriptomics reveal an association of cellular aging with MHC class I upregulation.Mol Metab. 2024 Sep;87:101990. doi: 10.1016/j.molmet.2024.101990. Epub 2024 Jul 14. Mol Metab. 2024. PMID: 39009220 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

