Interplay between Pds5 and Rec8 in regulating chromosome axis length and crossover frequency
- PMID: 33712462
- PMCID: PMC7954452
- DOI: 10.1126/sciadv.abe7920
Interplay between Pds5 and Rec8 in regulating chromosome axis length and crossover frequency
Abstract
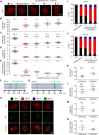
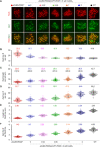
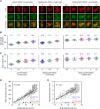
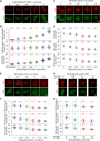
Meiotic chromosomes have a loop/axis architecture, with axis length determining crossover frequency. Meiosis-specific Pds5 depletion mutants have shorter chromosome axes and lower homologous chromosome pairing and recombination frequency. However, it is poorly understood how Pds5 coordinately regulates these processes. In this study, we show that only ~20% of wild-type level of Pds5 is required for homolog pairing and that higher levels of Pds5 dosage-dependently regulate axis length and crossover frequency. Moderate changes in Pds5 protein levels do not explicitly impair the basic recombination process. Further investigations show that Pds5 does not regulate chromosome axes by altering Rec8 abundance. Conversely, Rec8 regulates chromosome axis length by modulating Pds5. These findings highlight the important role of Pds5 in regulating meiosis and its relationship with Rec8 to regulate chromosome axis length and crossover frequency with implications for evolutionary adaptation.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

Similar articles
-
The ubiquitin-proteasome system regulates meiotic chromosome organization.Proc Natl Acad Sci U S A. 2022 Apr 26;119(17):e2106902119. doi: 10.1073/pnas.2106902119. Epub 2022 Apr 19. Proc Natl Acad Sci U S A. 2022. PMID: 35439061 Free PMC article.
-
Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe.Chromosoma. 2016 Jun;125(2):205-14. doi: 10.1007/s00412-015-0551-8. Epub 2015 Oct 28. Chromosoma. 2016. PMID: 26511279 Free PMC article.
-
ESA1 regulates meiotic chromosome axis and crossover frequency via acetylating histone H4.Nucleic Acids Res. 2021 Sep 20;49(16):9353-9373. doi: 10.1093/nar/gkab722. Nucleic Acids Res. 2021. PMID: 34417612 Free PMC article.
-
Crossover patterns under meiotic chromosome program.Asian J Androl. 2021 Nov-Dec;23(6):562-571. doi: 10.4103/aja.aja_86_20. Asian J Androl. 2021. PMID: 33533735 Free PMC article. Review.
-
The nature of meiotic chromosome dynamics and recombination in budding yeast.J Microbiol. 2019 Apr;57(4):221-231. doi: 10.1007/s12275-019-8541-9. Epub 2019 Jan 22. J Microbiol. 2019. PMID: 30671743 Review.
Cited by
-
Alterations in synaptonemal complex coding genes and human infertility.Int J Biol Sci. 2022 Feb 21;18(5):1933-1943. doi: 10.7150/ijbs.67843. eCollection 2022. Int J Biol Sci. 2022. PMID: 35342360 Free PMC article. Review.
-
The meiotic cohesin subunit REC8 contributes to multigenic adaptive evolution of autopolyploid meiosis in Arabidopsis arenosa.PLoS Genet. 2022 Jul 13;18(7):e1010304. doi: 10.1371/journal.pgen.1010304. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35830475 Free PMC article.
-
Per-nucleus crossover covariation is regulated by chromosome organization.iScience. 2022 Mar 18;25(4):104115. doi: 10.1016/j.isci.2022.104115. eCollection 2022 Apr 15. iScience. 2022. PMID: 35391833 Free PMC article.
-
All Ways Lead to Rome-Meiotic Stabilization Can Take Many Routes in Nascent Polyploid Plants.Genes (Basel). 2022 Jan 15;13(1):147. doi: 10.3390/genes13010147. Genes (Basel). 2022. PMID: 35052487 Free PMC article. Review.
-
The ubiquitin-proteasome system regulates meiotic chromosome organization.Proc Natl Acad Sci U S A. 2022 Apr 26;119(17):e2106902119. doi: 10.1073/pnas.2106902119. Epub 2022 Apr 19. Proc Natl Acad Sci U S A. 2022. PMID: 35439061 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

