Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions
- PMID: 33712587
- PMCID: PMC7954844
- DOI: 10.1038/s41467-021-21361-7
Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions
Abstract
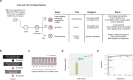
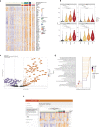
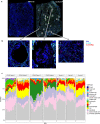
In less than nine months, the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) killed over a million people, including >25,000 in New York City (NYC) alone. The COVID-19 pandemic caused by SARS-CoV-2 highlights clinical needs to detect infection, track strain evolution, and identify biomarkers of disease course. To address these challenges, we designed a fast (30-minute) colorimetric test (LAMP) for SARS-CoV-2 infection from naso/oropharyngeal swabs and a large-scale shotgun metatranscriptomics platform (total-RNA-seq) for host, viral, and microbial profiling. We applied these methods to clinical specimens gathered from 669 patients in New York City during the first two months of the outbreak, yielding a broad molecular portrait of the emerging COVID-19 disease. We find significant enrichment of a NYC-distinctive clade of the virus (20C), as well as host responses in interferon, ACE, hematological, and olfaction pathways. In addition, we use 50,821 patient records to find that renin-angiotensin-aldosterone system inhibitors have a protective effect for severe COVID-19 outcomes, unlike similar drugs. Finally, spatial transcriptomic data from COVID-19 patient autopsy tissues reveal distinct ACE2 expression loci, with macrophage and neutrophil infiltration in the lungs. These findings can inform public health and may help develop and drive SARS-CoV-2 diagnostic, prevention, and treatment strategies.
Conflict of interest statement
N.T. and B.W.L. are employees at New England Biolabs. R.E.S. is on the scientific advisory board of Miromatrix Inc. Biotia employees and advisors include M.C.R., D.N.S., J.B., H.W., C.E.M., and N.O. D.B. is a co-founder of Poppy Inc; A.F and C.S.C are employees of DNAnexus. Authors not listed here do not have competing interests.
Figures



Update of
-
Shotgun Transcriptome and Isothermal Profiling of SARS-CoV-2 Infection Reveals Unique Host Responses, Viral Diversification, and Drug Interactions.bioRxiv [Preprint]. 2020 May 1:2020.04.20.048066. doi: 10.1101/2020.04.20.048066. bioRxiv. 2020. Update in: Nat Commun. 2021 Mar 12;12(1):1660. doi: 10.1038/s41467-021-21361-7. PMID: 32511352 Free PMC article. Updated. Preprint.
References
-
- Zhao, Z., Sokhansanj, B. A. & Rosen, G. Characterizing geographical and temporal dynamics of novel coronavirus SARS-CoV-2 using informative subytype markers. BioRxiv 10.1101/2020.04.07.030759v3 (2020).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T15 LM007079/LM/NLM NIH HHS/United States
- TL1 TR002386/TR/NCATS NIH HHS/United States
- U19 AI144297/AI/NIAID NIH HHS/United States
- R35 GM138152/GM/NIGMS NIH HHS/United States
- UL1 TR002384/TR/NCATS NIH HHS/United States
- R01 DK121072/DK/NIDDK NIH HHS/United States
- R35 GM131905/GM/NIGMS NIH HHS/United States
- UL1 TR000457/TR/NCATS NIH HHS/United States
- U01 DA053941/DA/NIDA NIH HHS/United States
- R01 MH117406/MH/NIMH NIH HHS/United States
- R25 EB020393/EB/NIBIB NIH HHS/United States
- R01 AI151059/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

