A novel allele of SIR2 reveals a heritable intermediate state of gene silencing
- PMID: 33713126
- PMCID: PMC8128386
- DOI: 10.1093/genetics/iyab041
A novel allele of SIR2 reveals a heritable intermediate state of gene silencing
Abstract
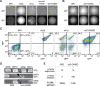
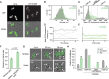
Genetic information acquires additional meaning through epigenetic regulation, the process by which genetically identical cells can exhibit heritable differences in gene expression and phenotype. Inheritance of epigenetic information is a critical step in maintaining cellular identity and organismal health. In Saccharomyces cerevisiae, one form of epigenetic regulation is the transcriptional silencing of two mating-type loci, HML and HMR, by the SIR-protein complex. To focus on the epigenetic dimension of this gene regulation, we conducted a forward mutagenesis screen to identify mutants exhibiting an epigenetic or metastable silencing defect. We utilized fluorescent reporters at HML and HMR, and screened yeast colonies for epigenetic silencing defects. We uncovered numerous independent sir1 alleles, a gene known to be required for stable epigenetic inheritance. More interestingly, we recovered a missense mutation within SIR2, which encodes a highly conserved histone deacetylase. In contrast to sir1Δ, which exhibits states that are either fully silenced or fully expressed, this sir2 allele exhibited heritable states that were either fully silenced or expressed at an intermediate level. The heritable nature of this unique silencing defect was influenced by, but not completely dependent on, changes in rDNA copy number. Therefore, this study revealed a heritable state of intermediate silencing and linked this state to a central silencing factor, Sir2.
Keywords: Sir2; epigenetic inheritance; transcriptional silencing.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Bähler J, Wu J-Q, Longtine MS, Shah NG, Mckenzie A III, et al.1998. Heterologous modules for efficient and versatile PCR‐based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. - PubMed
-
- Chien CT, Buck S, Sternglanz R, Shore D.. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531–541. - PubMed
-
- Dhillon N, Kamakaka RT.. 2000. A Histone Variant, Htz1p, and a Sir1p-like Protein, Esc2p, Mediate Silencing at HMR. Mol Cell 6:769–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

