A COVID-19 risk score combining chest CT radiomics and clinical characteristics to differentiate COVID-19 pneumonia from other viral pneumonias
- PMID: 33713401
- PMCID: PMC8064216
- DOI: 10.18632/aging.202735
A COVID-19 risk score combining chest CT radiomics and clinical characteristics to differentiate COVID-19 pneumonia from other viral pneumonias
Abstract
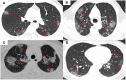
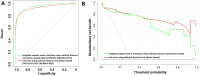
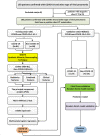
With the continued transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) throughout the world, identification of highly suspected COVID-19 patients remains an urgent priority. In this study, we developed and validated COVID-19 risk scores to identify patients with COVID-19. In this study, for patient-wise analysis, three signatures, including the risk score using radiomic features only, the risk score using clinical factors only, and the risk score combining radiomic features and clinical variables, show an excellent performance in differentiating COVID-19 from other viral-induced pneumonias in the validation set. For lesion-wise analysis, the risk score using three radiomic features only also achieved an excellent AUC value. In contrast, the performance of 130 radiologists based on the chest CT images alone without the clinical characteristics included was moderate as compared to the risk scores developed. The risk scores depicting the correlation of CT radiomics and clinical factors with COVID-19 could be used to accurately identify patients with COVID-19, which would have clinically translatable diagnostic and therapeutic implications from a precision medicine perspective.
Keywords: COVID-19; chest CT; coronavirus disease 2019; nomogram; radiomics; severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
Figures




References
-
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020; 395:1054–62. 10.1016/S0140-6736(20)30566-3 - DOI - PMC - PubMed
-
- Johns Hopkins University of Medicine. Coronavirus Resource Center. https://coronavirus.jhu.edu/
-
- Nevens D, Billiet C, Weytjens R, Joye I, Machiels M, Vermylen A, Chiari I, Bauwens W, Vermeulen P, Dirix L, Huget P, Verellen D, Dirix P, Meijnders P. The use of simulation-CT’s as a coronavirus disease 2019 screening tool during the severe acute respiratory syndrome coronavirus 2 pandemic. Radiother Oncol. 2020; 151:17–19. 10.1016/j.radonc.2020.07.006 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

