Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system
- PMID: 33713620
- PMCID: PMC7923941
- DOI: 10.1016/j.cell.2021.02.053
Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system
Erratum in
-
Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system.Cell. 2023 Nov 22;186(24):5428-5432. doi: 10.1016/j.cell.2023.10.008. Cell. 2023. PMID: 37995658 Free PMC article. No abstract available.
Abstract
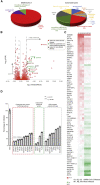
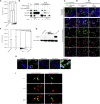
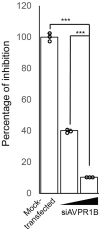
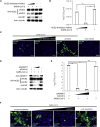
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause acute respiratory disease and multiorgan failure. Finding human host factors that are essential for SARS-CoV-2 infection could facilitate the formulation of treatment strategies. Using a human kidney cell line-HK-2-that is highly susceptible to SARS-CoV-2, we performed a genome-wide RNAi screen and identified virus dependency factors (VDFs), which play regulatory roles in biological pathways linked to clinical manifestations of SARS-CoV-2 infection. We found a role for a secretory form of SARS-CoV-2 receptor, soluble angiotensin converting enzyme 2 (sACE2), in SARS-CoV-2 infection. Further investigation revealed that SARS-CoV-2 exploits receptor-mediated endocytosis through interaction between its spike with sACE2 or sACE2-vasopressin via AT1 or AVPR1B, respectively. Our identification of VDFs and the regulatory effect of sACE2 on SARS-CoV-2 infection shed insight into pathogenesis and cell entry mechanisms of SARS-CoV-2 as well as potential treatment strategies for COVID-19.
Keywords: ACE2; COVID-19; RNAi; SARS-CoV-2; sACE2; vasopressin; virus dependency factor.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. The other authors declare no competing interests.
Figures
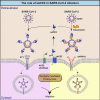
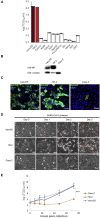
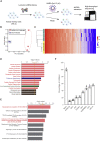
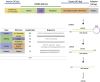

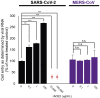


Comment in
-
Soluble ACE2 in SARS-CoV-2 infection.Nat Rev Nephrol. 2021 May;17(5):297. doi: 10.1038/s41581-021-00422-6. Nat Rev Nephrol. 2021. PMID: 33758362 Free PMC article. No abstract available.
-
Vasopressin infusion in COVID-19 critical illness is not associated with impaired viral clearance: a pilot study.Br J Anaesth. 2021 Oct;127(4):e146-e148. doi: 10.1016/j.bja.2021.07.005. Epub 2021 Jul 20. Br J Anaesth. 2021. PMID: 34399981 Free PMC article. No abstract available.
-
Evidence in favor of the essentiality of human cell membrane-bound ACE2 and against soluble ACE2 for SARS-CoV-2 infectivity.Cell. 2022 May 26;185(11):1837-1839. doi: 10.1016/j.cell.2022.05.004. Cell. 2022. PMID: 35623327 Free PMC article. No abstract available.
-
Response to Evidence in favor of the essentiality of human cell membrane-bound ACE2 and against soluble ACE2 for SARS-CoV-2 infectivity.Cell. 2022 May 26;185(11):1840-1841. doi: 10.1016/j.cell.2022.05.005. Cell. 2022. PMID: 35623328 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

