A Multimodality Machine Learning Approach to Differentiate Severe and Nonsevere COVID-19: Model Development and Validation
- PMID: 33714935
- PMCID: PMC8030658
- DOI: 10.2196/23948
A Multimodality Machine Learning Approach to Differentiate Severe and Nonsevere COVID-19: Model Development and Validation
Abstract
Background: Effectively and efficiently diagnosing patients who have COVID-19 with the accurate clinical type of the disease is essential to achieve optimal outcomes for the patients as well as to reduce the risk of overloading the health care system. Currently, severe and nonsevere COVID-19 types are differentiated by only a few features, which do not comprehensively characterize the complicated pathological, physiological, and immunological responses to SARS-CoV-2 infection in the different disease types. In addition, these type-defining features may not be readily testable at the time of diagnosis.
Objective: In this study, we aimed to use a machine learning approach to understand COVID-19 more comprehensively, accurately differentiate severe and nonsevere COVID-19 clinical types based on multiple medical features, and provide reliable predictions of the clinical type of the disease.
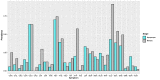
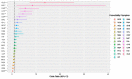
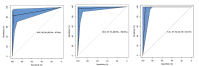
Methods: For this study, we recruited 214 confirmed patients with nonsevere COVID-19 and 148 patients with severe COVID-19. The clinical characteristics (26 features) and laboratory test results (26 features) upon admission were acquired as two input modalities. Exploratory analyses demonstrated that these features differed substantially between two clinical types. Machine learning random forest models based on all the features in each modality as well as on the top 5 features in each modality combined were developed and validated to differentiate COVID-19 clinical types.
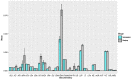
Results: Using clinical and laboratory results independently as input, the random forest models achieved >90% and >95% predictive accuracy, respectively. The importance scores of the input features were further evaluated, and the top 5 features from each modality were identified (age, hypertension, cardiovascular disease, gender, and diabetes for the clinical features modality, and dimerized plasmin fragment D, high sensitivity troponin I, absolute neutrophil count, interleukin 6, and lactate dehydrogenase for the laboratory testing modality, in descending order). Using these top 10 multimodal features as the only input instead of all 52 features combined, the random forest model was able to achieve 97% predictive accuracy.
Conclusions: Our findings shed light on how the human body reacts to SARS-CoV-2 infection as a unit and provide insights on effectively evaluating the disease severity of patients with COVID-19 based on more common medical features when gold standard features are not available. We suggest that clinical information can be used as an initial screening tool for self-evaluation and triage, while laboratory test results should be applied when accuracy is the priority.
Keywords: COVID-19; classification; clinical type; decision support; diagnosis; machine learning; multimodality; prediction; reliable.
©Yuanfang Chen, Liu Ouyang, Forrest S Bao, Qian Li, Lei Han, Hengdong Zhang, Baoli Zhu, Yaorong Ge, Patrick Robinson, Ming Xu, Jie Liu, Shi Chen. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 07.04.2021.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
References
-
- Weekly epidemiological update - 12 January 2021. World Health Organization. 2021. Jan 12, [2021-03-18]. https://www.who.int/publications/m/item/weekly-epidemiological-update---....
-
- Wölfel Roman, Corman Victor M, Guggemos Wolfgang, Seilmaier Michael, Zange Sabine, Müller Marcel A, Niemeyer Daniela, Jones Terry C, Vollmar Patrick, Rothe Camilla, Hoelscher Michael, Bleicker Tobias, Brünink Sebastian, Schneider Julia, Ehmann Rosina, Zwirglmaier Katrin, Drosten Christian, Wendtner Clemens. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 May;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. doi: 10.1038/s41586-020-2196-x. - DOI - PubMed
-
- Luo Yi, Trevathan Edwin, Qian Zhengmin, Li Yirong, Li Jin, Xiao Wei, Tu Ning, Zeng Zhikun, Mo Pingzheng, Xiong Yong, Ye Guangming. Asymptomatic SARS-CoV-2 Infection in Household Contacts of a Healthcare Provider, Wuhan, China. Emerg Infect Dis. 2020 Aug;26(8):1930–1933. doi: 10.3201/eid2608.201016. doi: 10.3201/eid2608.201016. - DOI - PMC - PubMed
-
- Ye F, Xu S, Rong Z, Xu R, Liu X, Deng P, Liu H, Xu X. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020 May;94:133–138. doi: 10.1016/j.ijid.2020.03.042. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(20)30174-0 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

