Exploring hTERT promoter methylation in cutaneous T-cell lymphomas
- PMID: 33715271
- PMCID: PMC9067155
- DOI: 10.1002/1878-0261.12946
Exploring hTERT promoter methylation in cutaneous T-cell lymphomas
Abstract
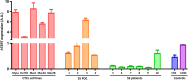
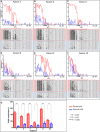
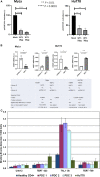
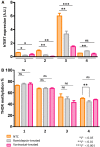
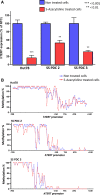
Cutaneous T-cell lymphomas (CTCLs) are telomerase-positive tumors expressing hTERT, although neither gene rearrangement/amplification nor promoter hotspot mutations could explain the hTERT re-expression. As the hTERT promoter is rich in CpG, we investigated the contribution of epigenetic mechanisms in its re-expression. We analyzed hTERT promoter methylation status in CTCL cells compared with healthy cells. Gene-specific methylation analyses revealed a common methylation pattern exclusively in tumor cells. This methylation pattern encompassed a hypermethylated distal region from -650 to -150 bp and a hypomethylated proximal region from -150 to +150 bp. Interestingly, the hypermethylated region matches with the recently named TERT hypermethylated oncogenic region (THOR). THOR has been associated with telomerase reactivation in many cancers, but it has so far not been reported in cutaneous lymphomas. Additionally, we assessed the effect of THOR on two histone deacetylase inhibitors (HDACi), romidepsin and vorinostat, both approved for CTCL treatment and a DNA methyltransferase inhibitor (DNMTi) 5-azacytidine, unapproved for CTCL. Contrary to our expectations, the findings reported herein revealed that THOR methylation is relatively stable under these epigenetic drugs' pressure, whereas these drugs reduced the hTERT gene expression.
Keywords: DNA methylation; DNMTi; HDACi; TERT; cutaneous T-cell lymphomas; telomerase.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz‐Perez JL, Duncan LM et al. (2005) WHO‐EORTC classification for cutaneous lymphomas. Blood 105, 3768–3785. - PubMed
-
- Willemze R, Hodak E, Zinzani PL, Specht L, Ladetto M & ESMO Guidelines Committee (2018) Primary cutaneous lymphomas: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann Oncol 29 (Suppl 4), iv30–iv40. - PubMed
-
- Hristov AC, Tejasvi T & Wilcox RA (2019) Mycosis fungoides and Sézary syndrome: 2019 update on diagnosis, risk‐stratification, and management. Am J Hematol 94, 1027–1041. - PubMed
-
- Finkel T, Serrano M & Blasco MA (2007) The common biology of cancer and ageing. Nature 448, 767–774. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

