The Mafb cleft-associated variant H131Q is not required for palatogenesis in the mouse
- PMID: 33715275
- PMCID: PMC9266196
- DOI: 10.1002/dvdy.327
The Mafb cleft-associated variant H131Q is not required for palatogenesis in the mouse
Abstract
Background: Orofacial clefts (OFCs) are common birth defects with complex etiology. Genome wide association studies for OFC have identified SNPs in and near MAFB. MAFB is a transcription factor critical for structural development of digits, kidneys, skin, and brain. MAFB is also expressed in the craniofacial region. Previous sequencing of MAFB in a Filipino population revealed a novel missense variant significantly associated with an increased risk for OFC. This MAFB variant, leading to the amino acid change H131Q, was knocked into the mouse Mafb, resulting in the MafbH131Q allele. The MafbH131Q construct was engineered to allow for deletion of Mafb ("Mafbdel ").
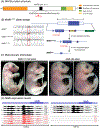
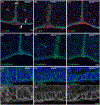
Results: Mafbdel/del animals died shortly after birth. Conversely, MafbH131Q/H131Q mice survived into adulthood at Mendelian ratios. Mafbdel/del and MafbH131Q/H131Q heads exhibited normal macroscopic and histological appearance at all embryonic time points evaluated. The periderm was intact based on expression of keratin 6, p63, and E-cadherin. Despite no effect on craniofacial morphogenesis, H131Q inhibited the Mafb-dependent promoter activation of Arhgap29 in palatal mesenchymal, but not ectodermal-derived epithelial cells in a luciferase assay.
Conclusions: Mafb is dispensable for murine palatogenesis in vivo, and the cleft-associated variant H131Q, despite its lack of morphogenic effect, altered the expression of Arhgap29 in a cell-dependent context.
Keywords: Mafb; craniofacial; development; mouse; mutation.
© 2021 American Association of Anatomists.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

