Scattered Light Imaging: Resolving the substructure of nerve fiber crossings in whole brain sections with micrometer resolution
- PMID: 33716156
- PMCID: PMC8204277
- DOI: 10.1016/j.neuroimage.2021.117952
Scattered Light Imaging: Resolving the substructure of nerve fiber crossings in whole brain sections with micrometer resolution
Abstract
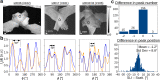
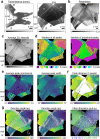
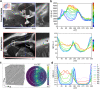
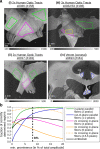
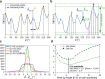
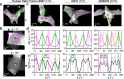
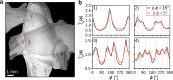
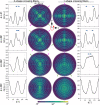
For developing a detailed network model of the brain based on image reconstructions, it is necessary to spatially resolve crossing nerve fibers. The accuracy hereby depends on many factors, including the spatial resolution of the imaging technique. 3D Polarized Light Imaging (3D-PLI) allows the three-dimensional reconstruction of nerve fiber tracts in whole brain sections with micrometer in-plane resolution, but leaves uncertainties in pixels containing crossing fibers. Here we introduce Scattered Light Imaging (SLI) to resolve the substructure of nerve fiber crossings. The measurement is performed on the same unstained histological brain sections as in 3D-PLI. By illuminating the brain sections from different angles and measuring the transmitted (scattered) light under normal incidence, light intensity profiles are obtained that are characteristic for the underlying brain tissue structure. We have developed a fully automated evaluation of the intensity profiles, allowing the user to extract various characteristics, like the individual directions of in-plane crossing nerve fibers, for each image pixel at once. We validate the reconstructed nerve fiber directions against results from previous simulation studies, scatterometry measurements, and fiber directions obtained from 3D-PLI. We demonstrate in different brain samples (human optic tracts, vervet monkey brain, rat brain) that the 2D fiber directions can be reliably reconstructed for up to three crossing nerve fiber bundles in each image pixel with an in-plane resolution of up to 6.5 μm. We show that SLI also yields reliable fiber directions in brain regions with low 3D-PLI signals coming from regions with a low density of myelinated nerve fibers or out-of-plane fibers. This makes Scattered Light Imaging a promising new imaging technique, providing crucial information about the organization of crossing nerve fibers in the brain.
Keywords: Brain structure; Connectivity; Light scattering; Nerve fiber crossings; White matter.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures









Similar articles
-
Scattering polarimetry enables correlative nerve fiber imaging and multimodal analysis.Sci Rep. 2025 May 27;15(1):18493. doi: 10.1038/s41598-025-02762-w. Sci Rep. 2025. PMID: 40425688 Free PMC article.
-
Scatterometry Measurements With Scattered Light Imaging Enable New Insights Into the Nerve Fiber Architecture of the Brain.Front Neuroanat. 2021 Nov 29;15:767223. doi: 10.3389/fnana.2021.767223. eCollection 2021. Front Neuroanat. 2021. PMID: 34912194 Free PMC article.
-
Imaging crossing fibers in mouse, pig, monkey, and human brain using small-angle X-ray scattering.Acta Biomater. 2023 Jul 1;164:317-331. doi: 10.1016/j.actbio.2023.04.029. Epub 2023 Apr 23. Acta Biomater. 2023. PMID: 37098400 Free PMC article.
-
Computational approaches for the reconstruction of optic nerve fibers along the visual pathway from medical images: a comprehensive review.Front Neurosci. 2023 May 26;17:1191999. doi: 10.3389/fnins.2023.1191999. eCollection 2023. Front Neurosci. 2023. PMID: 37304011 Free PMC article. Review.
-
Differential dynamic microscopy for the characterisation of motility in biological systems.Phys Chem Chem Phys. 2022 Sep 14;24(35):20616-20623. doi: 10.1039/d2cp02034c. Phys Chem Chem Phys. 2022. PMID: 36048134 Review.
Cited by
-
Evaluating backscattering polarized light imaging microstructural mapping capabilities through neural tissue and analogous phantom imaging.J Biomed Opt. 2024 May;29(5):052914. doi: 10.1117/1.JBO.29.5.052914. Epub 2023 Dec 5. J Biomed Opt. 2024. PMID: 38077501 Free PMC article.
-
Editorial: The human brain multiscale imaging challenge.Front Neuroanat. 2022 Oct 27;16:1060405. doi: 10.3389/fnana.2022.1060405. eCollection 2022. Front Neuroanat. 2022. PMID: 36387997 Free PMC article. No abstract available.
-
Scattering polarimetry enables correlative nerve fiber imaging and multimodal analysis.Sci Rep. 2025 May 27;15(1):18493. doi: 10.1038/s41598-025-02762-w. Sci Rep. 2025. PMID: 40425688 Free PMC article.
-
Using light and X-ray scattering to untangle complex neuronal orientations and validate diffusion MRI.Elife. 2023 May 11;12:e84024. doi: 10.7554/eLife.84024. Elife. 2023. PMID: 37166005 Free PMC article.
-
Micron-resolution fiber mapping in histology independent of sample preparation.bioRxiv [Preprint]. 2025 Mar 14:2024.03.26.586745. doi: 10.1101/2024.03.26.586745. bioRxiv. 2025. PMID: 38585744 Free PMC article. Preprint.
References
-
- Aboitiz F., Ide A., Olivares R. Corpus Callosum Morphology in Relation to Cerebral Asymmetries in the Postmortem Human. In: Zaidel E., Iacoboni M., editors. The Parallel Brain: The Cognitive Neuroscience of the Corpus Callosum. Massachusetts Institute of Technology; 2003. pp. 33–46.
-
- Axer M., Grässel D., Kleiner M., Dammers J., Dickscheid T., Reckfort J., Hütz T., Eiben B., Pietrzyk U., Zilles K., Amunts K. High-resolution fiber tract reconstruction in the human brain by means of three-dimensional polarized light imaging. Front. Neuroinform. 2011;5(34):1–13. doi: 10.3389/fninf.2011.00034. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

