Genomic Characterization of Two Shiga Toxin-Converting Bacteriophages Induced From Environmental Shiga Toxin-Producing Escherichia coli
- PMID: 33716997
- PMCID: PMC7946995
- DOI: 10.3389/fmicb.2021.587696
Genomic Characterization of Two Shiga Toxin-Converting Bacteriophages Induced From Environmental Shiga Toxin-Producing Escherichia coli
Abstract
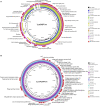
Shiga toxin (Stx), encoded by stx genes located in prophage sequences, is the major agent responsible for the pathogenicity of Shiga toxin-producing Escherichia coli (STEC) and is closely associated with the development of hemolytic uremic syndrome (HUS). Although numerous Stx prophage sequences have been reported as part of STEC bacterial genomes, the information about the genomic characterization of Stx-converting bacteriophages induced from STEC strains is relatively scarce. The objectives of this study were to genomically characterize two Stx-converting phages induced from environmental STEC strains and to evaluate their correlations with published Stx-converting phages and STEC strains of different origins. The Stx1-converting phage Lys8385Vzw and the Stx2-converting phage Lys19259Vzw were induced from E. coli O103:H11 (RM8385) and E. coli O157:H7 (RM19259), respectively. Whole-genome sequencing of these phages was conducted on a MiSeq sequencer for genomic characterization. Phylogenetic analysis and comparative genomics were performed to determine the correlations between these two Stx-converting phages, 13 reference Stx-converting phages, and 10 reference STEC genomes carrying closely related Stx prophages. Both Stx-converting phages Lys8385Vzw and Lys19259Vzw had double-stranded DNA, with genome sizes of 50,953 and 61,072 bp, respectively. Approximately 40% of the annotated coding DNA sequences with the predicted functions were likely associated with the fitness for both phages and their bacterial hosts. The whole-genome-based phylogenetic analysis of these two Stx-converting phages and 13 reference Stx-converting phages revealed that the 15 Stx-converting phages were divided into three distinct clusters, and those from E. coli O157:H7, in particular, were distributed in each cluster, demonstrating the high genomic diversity of these Stx-converting phages. The genomes of Stx-converting phage Lys8385Vzw and Lys19259Vzw shared a high-nucleotide similarity with the prophage sequences of the selected STEC isolates from the clinical and environmental origin. The findings demonstrate the genomic diversity of Stx-converting phages induced from different STEC strains and provide valuable insights into the dissemination of stx genes among E. coli population via the lysogenization of Stx-converting phages.
Keywords: Shiga toxin-producing Escherichia coli; Stx-converting bacteriophages; comparative genomics; genetic diversity; virulence gene transfer.
Copyright © 2021 Zhang, Liao, Salvador and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Allison H. E., Sergeant M. J., James C. E., Saunders J. R., Smith D. L., Sharp R. J., et al. (2003). Immunity profiles of wild-type and recombinant shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71 3409–3418. 10.1128/IAI.71.6.3409-3418.2003 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

