Sensing, Uptake and Catabolism of L-Phenylalanine During 2-Phenylethanol Biosynthesis via the Ehrlich Pathway in Saccharomyces cerevisiae
- PMID: 33717002
- PMCID: PMC7947893
- DOI: 10.3389/fmicb.2021.601963
Sensing, Uptake and Catabolism of L-Phenylalanine During 2-Phenylethanol Biosynthesis via the Ehrlich Pathway in Saccharomyces cerevisiae
Abstract
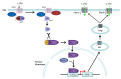
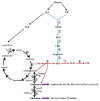
2-Phenylethanol (2-PE) is an important flavouring ingredient with a persistent rose-like odour, and it has been widely utilized in food, perfume, beverages, and medicine. Due to the potential existence of toxic byproducts in 2-PE resulting from chemical synthesis, the demand for "natural" 2-PE through biotransformation is increasing. L-Phenylalanine (L-Phe) is used as the precursor for the biosynthesis of 2-PE through the Ehrlich pathway by Saccharomyces cerevisiae. The regulation of L-Phe metabolism in S. cerevisiae is complicated and elaborate. We reviewed current progress on the signal transduction pathways of L-Phe sensing, uptake of extracellular L-Phe and 2-PE synthesis from L-Phe through the Ehrlich pathway. Moreover, the anticipated bottlenecks and future research directions for S. cerevisiae biosynthesis of 2-PE are discussed.
Keywords: 2-phenylethanol; Ehrlich pathway; Saccharomyces cerevisiae; sensing of L-phenylalanine; uptake of L-phenylalanine.
Copyright © 2021 Dai, Xia, Yang and Chen.
Conflict of interest statement
The authors declare that this study received funding from China Tobacco Corporation and Hubei tobacco company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Figures

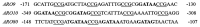
References
-
- Abdel-Sater F., Iraqui I., Urrestarazu A., André B. (2004). The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166, 1727–1739. 10.1534/genetics.166.4.1727, PMID: - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

