Low Bifidobacterium Abundance in the Lower Gut Microbiota Is Associated With Helicobacter pylori-Related Gastric Ulcer and Gastric Cancer
- PMID: 33717022
- PMCID: PMC7953064
- DOI: 10.3389/fmicb.2021.631140
Low Bifidobacterium Abundance in the Lower Gut Microbiota Is Associated With Helicobacter pylori-Related Gastric Ulcer and Gastric Cancer
Abstract
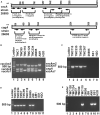
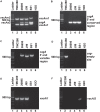
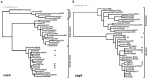
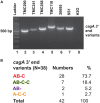
Helicobacter pylori infection in stomach leads to gastric cancer, gastric ulcer, and duodenal ulcer. More than 1 million people die each year due to these diseases, but why most H. pylori-infected individuals remain asymptomatic while a certain proportion develops such severe gastric diseases remained an enigma. Several studies indicated that gastric and intestinal microbiota may play a critical role in the development of the H. pylori-associated diseases. However, no specific microbe in the gastric or intestinal microbiota has been clearly linked to H. pylori infection and related gastric diseases. Here, we studied H. pylori infection, its virulence genes, the intestinal microbiota, and the clinical status of Trivandrum residents (N = 375) in southwestern India by standard H. pylori culture, PCR genotype, Sanger sequencing, and microbiome analyses using Illumina Miseq and Nanopore GridION. Our analyses revealed that gastric colonization by virulent H. pylori strains (vacAs1i1m1cagA+) is necessary but not sufficient for developing these diseases. Conversely, distinct microbial pools exist in the lower gut of the H. pylori-infected vs. H. pylori-non-infected individuals. Bifidobacterium (belonging to the phylum Actinobacteria) and Bacteroides (belonging to the phylum Bacteroidetes) were present in lower relative abundance for the H. pylori+ group than the H. pylori- group (p < 0.05). On the contrary, for the H. pylori+ group, genus Dialister (bacteria belonging to the phylum Firmicutes) and genus Prevotella (bacteria belonging to the phylum Bacteroidetes) were present in higher abundance compared to the H. pylori- group (p < 0.05). Notably, those who carried H. pylori in the stomach and had developed aggressive gastric diseases also had extremely low relative abundance (p < 0.05) of several Bifidobacterium species (e.g., B. adolescentis, B. longum) in the lower gut suggesting a protective role of Bifidobacterium. Our results show the link between lower gastrointestinal microbes and upper gastrointestinal diseases. Moreover, the results are important for developing effective probiotic and early prognosis of severe gastric diseases.
Keywords: Bifidobacterium; H. pylori; gastric ulcer and cancer; gut microbiome; probiotic.
Copyright © 2021 Devi, Devadas, George, Gandhimathi, Chouhan, Retnakumar, Alexander, Varghese, Dharmaseelan, Chandrika, Jissa, Das, Nair and Chattopadhyay.
Conflict of interest statement
AG was employed by the company Genotypic Technology Pvt. Ltd, Bengaluru, Karnataka, India. The NGS was outsourced to this company. Some of the analysis was done by AG upon request in the absence of any commercial or financial relationships. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Atherton J. C., Cao P., Peek R. M., Jr., Tummuru M. K., Blaser M. J., Cover T. L. (1995). Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270 17771–17777. 10.1074/jbc.270.30.17771 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

