Influenza B Virus Infection Is Enhanced Upon Heterotypic Co-infection With Influenza A Virus
- PMID: 33717023
- PMCID: PMC7947630
- DOI: 10.3389/fmicb.2021.631346
Influenza B Virus Infection Is Enhanced Upon Heterotypic Co-infection With Influenza A Virus
Abstract
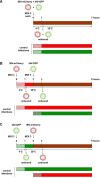
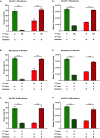
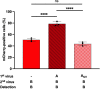
Homotypic co-infections with influenza viruses are described to increase genetic population diversity, to drive viral evolution and to allow genetic complementation. Less is known about heterotypic co-infections between influenza A (IAV) and influenza B (IBV) viruses. Previous publications showed that IAV replication was suppressed upon co-infection with IBV. However, the effect of heterotypic co-infections on IBV replication was not investigated. To do so, we produced by reverse genetics a pair of replication-competent recombinant IAV (A/WSN/33) and IBV (B/Brisbane/60/2008) expressing a GFP and mCherry fluorescent reporter, respectively. A549 cells were infected simultaneously or 1 h apart at a high MOI with IAV-GFP or IBV-mCherry and the fluorescence was measured at 6 h post-infection by flow cytometry. Unexpectedly, we observed that IBV-mCherry infection was enhanced upon co-infection with IAV-GFP, and more strongly so when IAV was added 1 h prior to IBV. The same effect was observed with wild-type viruses and with various strains of IAV. Using UV-inactivated IAV or type-specific antiviral compounds, we showed that the enhancing effect of IAV infection on IBV infection was dependent on transcription/replication of the IAV genome. Our results, taken with available data in the literature, support the hypothesis that the presence of IAV proteins can enhance IBV genome expression and/or complement IBV defective particles.
Keywords: co-infection; heterotypic; influenza B virus; influenza virus; viral interference.
Copyright © 2021 Malausse, van der Werf, Naffakh and Munier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Biquand E., Poirson J., Karim M., Declercq M., Malausse N., Cassonnet P., et al. (2017). Comparative profiling of ubiquitin proteasome system interplay with influenza A virus PB2 polymerase protein recapitulating virus evolution in humans. mSphere 2 00317–00330. 10.1128/mSphere.00330-17 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

