Effect of Low-Input Organic and Conventional Farming Systems on Maize Rhizosphere in Two Portuguese Open-Pollinated Varieties (OPV), "Pigarro" (Improved Landrace) and "SinPre" (a Composite Cross Population)
- PMID: 33717028
- PMCID: PMC7953162
- DOI: 10.3389/fmicb.2021.636009
Effect of Low-Input Organic and Conventional Farming Systems on Maize Rhizosphere in Two Portuguese Open-Pollinated Varieties (OPV), "Pigarro" (Improved Landrace) and "SinPre" (a Composite Cross Population)
Abstract
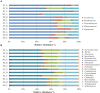
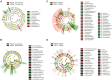
Maize is one of the most important crops worldwide and is the number one arable crop in Portugal. A transition from the conventional farming system to organic agriculture requires optimization of cultivars and management, the interaction of plant-soil rhizosphere microbiota being pivotal. The objectives of this study were to unravel the effect of population genotype and farming system on microbial communities in the rhizosphere of maize. Rhizosphere soil samples of two open-pollinated maize populations ("SinPre" and "Pigarro") cultivated under conventional and organic farming systems were taken during flowering and analyzed by next-generation sequencing (NGS). Phenological data were collected from the replicated field trial. A total of 266 fungi and 317 bacteria genera were identified in "SinPre" and "Pigarro" populations, of which 186 (69.9%) and 277 (87.4%) were shared among them. The microbiota of "Pigarro" showed a significant higher (P < 0.05) average abundance than the microbiota of "SinPre." The farming system had a statistically significant impact (P < 0.05) on the soil rhizosphere microbiota, and several fungal and bacterial taxa were found to be farming system-specific. The rhizosphere microbiota diversity in the organic farming system was higher than that in the conventional system for both varieties. The presence of arbuscular mycorrhizae (Glomeromycota) was mainly detected in the microbiota of the "SinPre" population under the organic farming systems and very rare under conventional systems. A detailed metagenome function prediction was performed. At the fungal level, pathotroph-saprotroph and pathotroph-symbiotroph lifestyles were modified by the farming system. For bacterial microbiota, the main functions altered by the farming system were membrane transport, transcription, translation, cell motility, and signal transduction. This study allowed identifying groups of microorganisms known for their role as plant growth-promoting rhizobacteria (PGPR) and with the capacity to improve crop tolerance for stress conditions, allowing to minimize the use of synthetic fertilizers and pesticides. Arbuscular mycorrhizae (phyla Glomeromycota) were among the most important functional groups in the fungal microbiota and Achromobacter, Burkholderia, Erwinia, Lysinibacillus, Paenibacillus, Pseudomonas, and Stenotrophomonas in the bacterial microbiota. In this perspective, the potential role of these microorganisms will be explored in future research.
Keywords: maize; microbiota; next-generation sequencing; open-pollinated populations; organic and conventional farming system; rhizosphere.
Copyright © 2021 Ares, Costa, Joaquim, Pintado, Santos, Messmer and Mendes-Moreira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Ahmed M. S., Sallam N. M., Mohamed A. A., Hassan M. H. (2013). Effect of mycorrhiza and biofertilisers on reducing the incidence of Fusarium root and pod rot diseases of peanut. Arch. Phytopathol. 46 868–881. 10.1080/03235408.2012.753707 - DOI
-
- Altieri M., Merrick L. (1987). In situ conservation of crop genetic resources through maintenance of traditional farming systems. Econ. Bot. 41 86–96. 10.1007/BF02859354 - DOI
-
- ANPROMIS (2019). Associação Nacional de Produtores de Milho e Sorgo. Available online at: http://www.anpromis.pt/o-milho.html (access August 13, 2019).
LinkOut - more resources
Full Text Sources
Other Literature Sources

