Chemotranscriptomic Profiling Defines Drug-Specific Signatures of the Glycopeptide Antibiotics Dalbavancin, Vancomycin and Chlorobiphenyl-Vancomycin in a VanB-Type-Resistant Streptomycete
- PMID: 33717038
- PMCID: PMC7947799
- DOI: 10.3389/fmicb.2021.641756
Chemotranscriptomic Profiling Defines Drug-Specific Signatures of the Glycopeptide Antibiotics Dalbavancin, Vancomycin and Chlorobiphenyl-Vancomycin in a VanB-Type-Resistant Streptomycete
Abstract
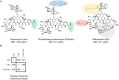
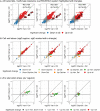
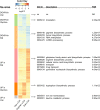
Dalbavancin, vancomycin and chlorobiphenyl-vancomycin share a high degree of structural similarity and the same primary mode of drug action. All inhibit bacterial cell wall biosynthesis through complexation with intermediates in peptidoglycan biosynthesis mediated via interaction with peptidyl-d-alanyl-d-alanine (d-Ala-d-Ala) residues present at the termini of the intermediates. VanB-type glycopeptide resistance in bacteria encodes an inducible reprogramming of bacterial cell wall biosynthesis that generates precursors terminating with d-alanyl-d-lactate (d-Ala-d-Lac). This system in Streptomyces coelicolor confers protection against the natural product vancomycin but not dalbavancin or chlorobiphenyl-vancomycin, which are semi-synthetic derivatives and fail to sufficiently activate the inducible VanB-type sensory response. We used transcriptome profiling by RNAseq to identify the gene expression signatures elucidated in S. coelicolor in response to the three different glycopeptide compounds. An integrated comparison of the results defines both the contribution of the VanB resistance system to the control of changes in gene transcription and the impact at the transcriptional level of the structural diversity present in the glycopeptide antibiotics used. Dalbavancin induces markedly more extensive changes in the expression of genes required for transport processes, RNA methylation, haem biosynthesis and the biosynthesis of the amino acids arginine and glutamine. Chlorobiphenyl-vancomycin exhibits specific effects on tryptophan and calcium-dependent antibiotic biosynthesis and has a stronger repressive effect on translation. Vancomycin predictably has a uniquely strong effect on the genes controlled by the VanB resistance system and also impacts metal ion homeostasis and leucine biosynthesis. Leaderless gene transcription is disfavoured in the core transcriptional up- and down-regulation taking place in response to all the glycopeptide antibiotics, while HrdB-dependent transcripts are favoured in the down-regulated group. This study illustrates the biological impact of peripheral changes to glycopeptide antibiotic structure and could inform the design of future semi-synthetic glycopeptide derivatives.
Keywords: Streptomyces; antibiotic; chemotranscriptomics; dalbavancin; glycopeptide; resistance; vancomycin.
Copyright © 2021 Hesketh, Bucca, Smith and Hong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andrews S. (2010). fastqc: A Quality Control Tool for High Throughput Sequence Data (v.0.11.8). Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Boshoff H. I., Myers T. G., Copp B. R., McNeil M. R., Wilson M. A., Barry C. E., III (2004). The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279 40174–40184. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

