Comparative Genomics of Emerging Lineages and Mobile Resistomes of Contemporary Broiler Strains of Salmonella Infantis and E. coli
- PMID: 33717039
- PMCID: PMC7947892
- DOI: 10.3389/fmicb.2021.642125
Comparative Genomics of Emerging Lineages and Mobile Resistomes of Contemporary Broiler Strains of Salmonella Infantis and E. coli
Abstract
Introduction: Commensal and pathogenic strains of multidrug-resistant (MDR) Escherichia coli and non-typhoid strains of Salmonella represent a growing foodborne threat from foods of poultry origin. MDR strains of Salmonella Infantis and E. coli are frequently isolated from broiler chicks and the simultaneous presence of these two enteric bacterial species would potentially allow the exchange of mobile resistance determinants.
Objectives: In order to understand possible genomic relations and to obtain a first insight into the potential interplay of resistance genes between enteric bacteria, we compared genomic diversity and mobile resistomes of S. Infantis and E. coli from broiler sources.
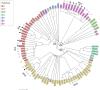
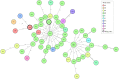
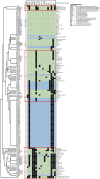
Results: The core genome MLST analysis of 56 S. Infantis and 90 E. coli contemporary strains revealed a high genomic heterogeneity of broiler E. coli. It also allowed the first insight into the genomic diversity of the MDR clone B2 of S. Infantis, which is endemic in Hungary. We also identified new MDR lineages for S. Infantis (ST7081 and ST7082) and for E. coli (ST8702 and ST10088). Comparative analysis of antibiotic resistance genes and plasmid types revealed a relatively narrow interface between the mobile resistomes of E. coli and S. Infantis. The mobile resistance genes tet(A), aadA1, and sul1 were identified at an overall high prevalence in both species. This gene association is characteristic to the plasmid pSI54/04 of the epidemic clone B2 of S. Infantis. Simultaneous presence of these genes and of IncI plasmids of the same subtype in cohabitant caecal strains of E. coli and S. Infantis suggests an important role of these plasmid families in a possible interplay of resistance genes between S. Infantis and E. coli in broilers.
Conclusion: This is the first comparative genomic analysis of contemporary broiler strains of S. Infantis and E. coli. The diversity of mobile resistomes suggests that commensal E. coli could be potential reservoirs of resistance for S. Infantis, but so far only a few plasmid types and mobile resistance genes could be considered as potentially exchangeable between these two species. Among these, IncI1 plasmids could make the greatest contribution to the microevolution and genetic interaction between E. coli and S. Infantis.
Keywords: Escherichia coli; Salmonella Infantis; antibiotic resistance; core genome; plasmid; resistome.
Copyright © 2021 Szmolka, Wami and Dobrindt.
Conflict of interest statement
The authors declare that this study received funding from Pharma-Zentrale GmbH (Herdecke). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of the article, or the decision to submit it for publication.
Figures



Similar articles
-
Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia.Int J Food Microbiol. 2020 Apr 16;319:108497. doi: 10.1016/j.ijfoodmicro.2019.108497. Epub 2019 Dec 26. Int J Food Microbiol. 2020. PMID: 31927155
-
Emergence of Multidrug-Resistant Salmonella enterica Subspecies enterica Serovar Infantis of Multilocus Sequence Type 2283 in German Broiler Farms.Front Microbiol. 2020 Jul 17;11:1741. doi: 10.3389/fmicb.2020.01741. eCollection 2020. Front Microbiol. 2020. PMID: 32765483 Free PMC article.
-
Recent Occurrence and Rapid Spread of Multidrug-Resistant Salmonella Infantis in Broiler Flocks in Korea.Foodborne Pathog Dis. 2025 Feb 27. doi: 10.1089/fpd.2024.0162. Online ahead of print. Foodborne Pathog Dis. 2025. PMID: 40014431
-
Salmonella enterica Serovar Infantis in Broiler Chickens: A Systematic Review and Meta-Analysis.Animals (Basel). 2024 Nov 28;14(23):3453. doi: 10.3390/ani14233453. Animals (Basel). 2024. PMID: 39682418 Free PMC article. Review.
-
Multidrug resistant commensal Escherichia coli in animals and its impact for public health.Front Microbiol. 2013 Sep 3;4:258. doi: 10.3389/fmicb.2013.00258. Front Microbiol. 2013. PMID: 24027562 Free PMC article. Review.
Cited by
-
Comparative genomics reveals high genetic similarity among strains of Salmonella enterica serovar Infantis isolated from multiple sources in Brazil.PeerJ. 2024 May 20;12:e17306. doi: 10.7717/peerj.17306. eCollection 2024. PeerJ. 2024. PMID: 38784399 Free PMC article.
-
Emergence and Comparative Genome Analysis of Salmonella Ohio Strains from Brown Rats, Poultry, and Swine in Hungary.Int J Mol Sci. 2024 Aug 13;25(16):8820. doi: 10.3390/ijms25168820. Int J Mol Sci. 2024. PMID: 39201506 Free PMC article.
-
Resilient by Design: Environmental Stress Promotes Biofilm Formation and Multi-Resistance in Poultry-Associated Salmonella.Microorganisms. 2025 Aug 3;13(8):1812. doi: 10.3390/microorganisms13081812. Microorganisms. 2025. PMID: 40871316 Free PMC article.
-
Prevalence of efflux pump and heavy metal tolerance encoding genes among Salmonella enterica serovar Infantis strains from diverse sources in Brazil.PLoS One. 2022 Nov 22;17(11):e0277979. doi: 10.1371/journal.pone.0277979. eCollection 2022. PLoS One. 2022. PMID: 36413564 Free PMC article.
-
Emergence and Genomic Features of a mcr-1 Escherichia coli from Duck in Hungary.Antibiotics (Basel). 2023 Oct 7;12(10):1519. doi: 10.3390/antibiotics12101519. Antibiotics (Basel). 2023. PMID: 37887221 Free PMC article.
References
-
- Alba P., Leekitcharoenphon P., Carfora V., Amoruso R., Cordaro G., Matteo P. D., et al. (2020). Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 6:e000365. 10.1099/mgen.0.000365 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

