Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism
- PMID: 33717070
- PMCID: PMC7952875
- DOI: 10.3389/fimmu.2021.592447
Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism
Abstract
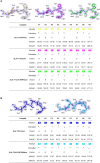
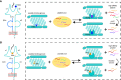
The micropolymorphism of major histocompatibility complex class I (MHC-I) can greatly alter the plasticity of peptide presentation, but elucidating the underlying mechanism remains a challenge. Here we investigated the impact of the micropolymorphism on peptide presentation of swine MHC-I (termed swine leukocyte antigen class I, SLA-I) molecules via immunopeptidomes that were determined by our newly developed random peptide library combined with the mass spectrometry (MS) de novo sequencing method (termed RPLD-MS) and the corresponding crystal structures. The immunopeptidomes of SLA-1*04:01, SLA-1*13:01, and their mutants showed that mutations of residues 156 and 99 could expand and narrow the ranges of peptides presented by SLA-I molecules, respectively. R156A mutation of SLA-1*04:01 altered the charge properties and enlarged the volume size of pocket D, which eliminated the harsh restriction to accommodate the third (P3) anchor residue of the peptide and expanded the peptide binding scope. Compared with 99Tyr of SLA-1*0401, 99Phe of SLA-1*13:01 could not form a conservative hydrogen bond with the backbone of the P3 residues, leading to fewer changes in the pocket properties but a significant decrease in quantitative of immunopeptidomes. This absent force could be compensated by the salt bridge formed by P1-E and 170Arg. These data illustrate two distinguishing manners that show how micropolymorphism alters the peptide-binding plasticity of SLA-I alleles, verifying the sensitivity and accuracy of the RPLD-MS method for determining the peptide binding characteristics of MHC-I in vitro and helping to more accurately predict and identify MHC-I restricted epitopes.
Keywords: RPLD–MS; crystal structure; immunopeptidome; micropolymorphism; swine MHC class I.
Copyright © 2021 Wei, Wang, Li, Li, Qu, Wang, Zou, Liang, Xia and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Peptidomes and Structures Illustrate How SLA-I Micropolymorphism Influences the Preference of Binding Peptide Length.Front Immunol. 2022 Feb 28;13:820881. doi: 10.3389/fimmu.2022.820881. eCollection 2022. Front Immunol. 2022. PMID: 35296092 Free PMC article.
-
Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.J Virol. 2022 Feb 23;96(4):e0137821. doi: 10.1128/JVI.01378-21. Epub 2021 Dec 1. J Virol. 2022. PMID: 34851145 Free PMC article.
-
A combined prediction strategy increases identification of peptides bound with high affinity and stability to porcine MHC class I molecules SLA-1*04:01, SLA-2*04:01, and SLA-3*04:01.Immunogenetics. 2016 Feb;68(2):157-65. doi: 10.1007/s00251-015-0883-9. Epub 2015 Nov 14. Immunogenetics. 2016. PMID: 26572135
-
Peptide binding motifs for MHC class I and II molecules.Curr Protoc Immunol. 2001 May;Appendix 1:Appendix 1I. doi: 10.1002/0471142735.ima01is36. Curr Protoc Immunol. 2001. PMID: 18432645 Review.
-
Molecular genetics of the swine major histocompatibility complex, the SLA complex.Dev Comp Immunol. 2009 Mar;33(3):362-74. doi: 10.1016/j.dci.2008.07.002. Epub 2008 Aug 27. Dev Comp Immunol. 2009. PMID: 18760302 Review.
Cited by
-
Peptidomes and Structures Illustrate How SLA-I Micropolymorphism Influences the Preference of Binding Peptide Length.Front Immunol. 2022 Feb 28;13:820881. doi: 10.3389/fimmu.2022.820881. eCollection 2022. Front Immunol. 2022. PMID: 35296092 Free PMC article.
-
New vistas unfold: Chicken MHC molecules reveal unexpected ways to present peptides to the immune system.Front Immunol. 2022 Jul 29;13:886672. doi: 10.3389/fimmu.2022.886672. eCollection 2022. Front Immunol. 2022. PMID: 35967451 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

