Spontaneously Resolved Atopic Dermatitis Shows Melanocyte and Immune Cell Activation Distinct From Healthy Control Skin
- PMID: 33717163
- PMCID: PMC7943477
- DOI: 10.3389/fimmu.2021.630892
Spontaneously Resolved Atopic Dermatitis Shows Melanocyte and Immune Cell Activation Distinct From Healthy Control Skin
Abstract
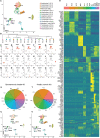
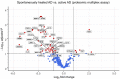
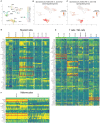
Atopic dermatitis (AD) typically starts in infancy or early childhood, showing spontaneous remission in a subset of patients, while others develop lifelong disease. Despite an increased understanding of AD, factors guiding its natural course are only insufficiently elucidated. We thus performed suction blistering in skin of adult patients with stable, spontaneous remission from previous moderate-to-severe AD during childhood. Samples were compared to healthy controls without personal or familial history of atopy, and to chronic, active AD lesions. Skin cells and tissue fluid obtained were used for single-cell RNA sequencing and proteomic multiplex assays, respectively. We found overall cell composition and proteomic profiles of spontaneously healed AD to be comparable to healthy control skin, without upregulation of typical AD activity markers (e.g., IL13, S100As, and KRT16). Among all cell types in spontaneously healed AD, melanocytes harbored the largest numbers of differentially expressed genes in comparison to healthy controls, with upregulation of potentially anti-inflammatory markers such as PLA2G7. Conventional T-cells also showed increases in regulatory markers, and a general skewing toward a more Th1-like phenotype. By contrast, gene expression of regulatory T-cells and keratinocytes was essentially indistinguishable from healthy skin. Melanocytes and conventional T-cells might thus contribute a specific regulatory milieu in spontaneously healed AD skin.
Keywords: atopic dermatitis; eczema; multiplex proteomics; single-cell RNA seq; spontaneous remission.
Copyright © 2021 Rindler, Krausgruber, Thaler, Alkon, Bangert, Kurz, Fortelny, Rojahn, Jonak, Griss, Bock and Brunner.
Conflict of interest statement
CBa is an employee of the Medical University of Vienna, and has received personal fees from Bayer, Mylan, LEO Pharma, Pfizer, Sanofi Genzyme, Eli Lilly, Novartis, Celgene, and AbbVie. CBa is an investigator for Novartis, Sanofi, Abbvie, Elli Lilly, and Galderma (grants paid to her institution). CJ is an employee of the Medical University of Vienna, and has received personal fees from LEO Pharma, Pfizer, Eli Lilly and Company, Novartis, Takeda, Mallinckrodt/Therakos, AbbVie, Janssen, and Almirall; and is an investigator for Eli Lilly and Company, Novartis, and 4SC (grants paid to her institution). PB is an employee of the Medical University of Vienna, and has received personal fees from LEO Pharma, Pfizer, Sanofi, Eli Lilly, Novartis, Celgene, UCB Pharma, Biotest, Boehringer Ingelheim, AbbVie, Amgen and Arena Pharmaceuticals. PB is an investigator for Novartis (grants paid to his institution). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis.J Allergy Clin Immunol. 2020 Nov;146(5):1056-1069. doi: 10.1016/j.jaci.2020.03.041. Epub 2020 Apr 25. J Allergy Clin Immunol. 2020. PMID: 32344053
-
Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities.J Allergy Clin Immunol. 2021 Apr;147(4):1369-1380. doi: 10.1016/j.jaci.2020.08.041. Epub 2020 Oct 1. J Allergy Clin Immunol. 2021. PMID: 33011244
-
Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis.J Allergy Clin Immunol. 2020 Jun;145(6):1615-1628. doi: 10.1016/j.jaci.2020.01.042. Epub 2020 Feb 7. J Allergy Clin Immunol. 2020. PMID: 32035984
-
Role of skin-homing t-cells in recurrent episodes of atopic dermatitis: a review.Front Immunol. 2025 Feb 18;16:1489277. doi: 10.3389/fimmu.2025.1489277. eCollection 2025. Front Immunol. 2025. PMID: 40040698 Free PMC article. Review.
-
Immune regulation in atopic dermatitis.Curr Opin Immunol. 2000 Dec;12(6):641-6. doi: 10.1016/s0952-7915(00)00156-4. Curr Opin Immunol. 2000. PMID: 11102766 Review.
Cited by
-
Applications of single-cell RNA sequencing in atopic dermatitis and psoriasis.Front Immunol. 2022 Nov 25;13:1038744. doi: 10.3389/fimmu.2022.1038744. eCollection 2022. Front Immunol. 2022. PMID: 36505405 Free PMC article. Review.
-
Single-Cell Transcriptional Analysis Deciphers the Inflammatory Response of Skin-Resident Stromal Cells.Front Surg. 2022 Jun 14;9:935107. doi: 10.3389/fsurg.2022.935107. eCollection 2022. Front Surg. 2022. PMID: 35774389 Free PMC article. Review.
-
Single-cell transcriptomics in human skin research: available technologies, technical considerations and disease applications.Exp Dermatol. 2022 May;31(5):655-673. doi: 10.1111/exd.14547. Epub 2022 Mar 4. Exp Dermatol. 2022. PMID: 35196402 Free PMC article. Review.
-
Generation of Schwann cell derived melanocytes from hPSCs identifies pro-metastatic factors in melanoma.bioRxiv [Preprint]. 2023 Mar 7:2023.03.06.531220. doi: 10.1101/2023.03.06.531220. bioRxiv. 2023. PMID: 36945537 Free PMC article. Preprint.
-
Comparison of CD30L and OX40L Reveals CD30L as a Promising Therapeutic Target in Atopic Dermatitis.Allergy. 2025 Feb;80(2):500-512. doi: 10.1111/all.16412. Epub 2024 Nov 26. Allergy. 2025. PMID: 39589186 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

