The phylogeny, phylogeography, and diversification history of the westernmost Asian cobra (Serpentes: Elapidae: Naja oxiana) in the Trans-Caspian region
- PMID: 33717439
- PMCID: PMC7920780
- DOI: 10.1002/ece3.7144
The phylogeny, phylogeography, and diversification history of the westernmost Asian cobra (Serpentes: Elapidae: Naja oxiana) in the Trans-Caspian region
Erratum in
-
Erratum.Ecol Evol. 2021 May 25;11(12):8394-8395. doi: 10.1002/ece3.7455. eCollection 2021 Jun. Ecol Evol. 2021. PMID: 34188894 Free PMC article.
Abstract
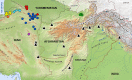
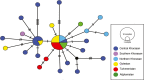
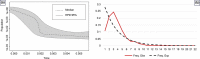
We conducted a comprehensive analysis of the phylogenetic, phylogeographic, and demographic relationships of Caspian cobra (Naja oxiana; Eichwald, 1831) populations based on a concatenated dataset of two mtDNA genes (cyt b and ND4) across the species' range in Iran, Afghanistan, and Turkmenistan, along with other members of Asian cobras (i.e., subgenus Naja Laurenti, 1768). Our results robustly supported that the Asiatic Naja are monophyletic, as previously suggested by other studies. Furthermore, N. kaouthia and N. sagittifera were recovered as sister taxa to each other, and in turn sister clades to N. oxiana. Our results also highlighted the existence of a single major evolutionary lineage for populations of N. oxiana in the Trans-Caspian region, suggesting a rapid expansion of this cobra from eastern to western Asia, coupled with a rapid range expansion from east of Iran toward the northeast. However, across the Iranian range of N. oxiana, subdivision of populations was not supported, and thus, a single evolutionary significant unit is proposed for inclusion in future conservation plans in this region.
Keywords: Naja oxiana; demographic history; phylogeny; phylogeography.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare to have no competing interests.
Figures





References
-
- Amato, G. D. (1991). Species hybridization and protection of endangered animals. Science, 253(5017), 250–252. - PubMed
-
- Arevalo, E. , Davis, S. K. , & Sites, J. W. (1994). Mitochondrial‐DNA sequence divergence and phylogenetic‐relationships among 8 chromosome races of the sceloporus–grammicus complex (Phrynosomatidae) in Central Mexico. Systematic Biology, 43(3), 387–418.
-
- Ashraf, M. R. , Nadeem, A. , Smith, E. N. , Javed, M. , Smart, U. , Yaqub, T. , & Hashmi, A. S. (2019). Molecular phylogenetics of black cobra (Naja naja) in Pakistan. Electronic Journal of Biotechnology, 42, 23–29. 10.1016/j.ejbt.2019.10.005 - DOI
-
- Avise, J. C. (2000). Speciation processes and extended genealogy. In Berendzen P. B., & Simons A. M. (Eds.), Phylogeography: The history and formation of specie (pp. 285–340). Harvard University Press.
-
- Ballard, J. W. O. , & Rand, D. M. (2005). The population biology of mitochondrial DNA and its phylogenetic implications. Annual Review of Ecology, Evolution, and Systematics, 36, 621–642.
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

