Drug target discovery by magnetic nanoparticles coupled mass spectrometry
- PMID: 33717618
- PMCID: PMC7930636
- DOI: 10.1016/j.jpha.2020.02.002
Drug target discovery by magnetic nanoparticles coupled mass spectrometry
Abstract
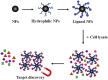
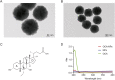
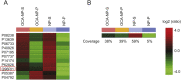
Drug target discovery is the basis of drug screening. It elucidates the cause of disease and the mechanism of drug action, which is the essential of drug innovation. Target discovery performed in biological systems is complicated as proteins are in low abundance and endogenous compounds may interfere with drug binding. Therefore, methods to track drug-target interactions in biological matrices are urgently required. In this work, a Fe3O4 nanoparticle-based approach was developed for drug-target screening in biofluids. A known ligand-protein complex was selected as a principle-to-proof example to validate the feasibility. After incubation in cell lysates, ligand-modified Fe3O4 nanoparticles bound to the target protein and formed complexes that were separated from the lysates by a magnet for further analysis. The large surface-to-volume ratio of the nanoparticles provides more active sites for the modification of chemical drugs. It enhances the opportunity for ligand-protein interactions, which is beneficial for capturing target proteins, especially for those with low abundance. Additionally, a one-step magnetic separation simplifies the pre-processing of ligand-protein complexes, so it effectively reduces the endogenous interference. Therefore, the present nanoparticle-based approach has the potential to be used for drug target screening in biological systems.
Keywords: Drug-target discovery; Mass spectrometry; Nanoparticle.
© 2020 Xi'an Jiaotong University. Production and hosting by Elsevier B.V.
Conflict of interest statement
Authors declare that there are no conflicts of interest.
Figures


References
-
- Betz U.A. How many genomics targets can a portfolio afford? Drug Discov. Today. 2005;10:1057–1063. - PubMed
-
- Miljenović T., Jia X., Lavrencic P. A non-uniform sampling approach enables studies of dilute and unstable proteins. J. Biomol. NMR. 2017;68:119–127. - PubMed
-
- Wyss D.F., Wang Y.S., Eaton H.L. Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors. Top. Curr. Chem. 2012;317:83–114. - PubMed
-
- Topf A., Franz P., Tsiavaliaris G. MicroScale Thermophoresis (MST) for studying actin polymerization kinetics. Biotechniques. 2017;63:187–190. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

